Genetic factors regulating Plasmodium falciparum gametocytogenesis identified by phenotypic screens
- PMID: 39730700
- PMCID: PMC11680961
- DOI: 10.1038/s41598-024-82133-z
Genetic factors regulating Plasmodium falciparum gametocytogenesis identified by phenotypic screens
Abstract
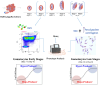
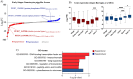
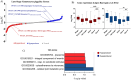
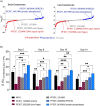
Successful transmission of Plasmodium falciparum from one person to another relies on the complete intraerythrocytic development of non-pathogenic sexual gametocytes infectious for anopheline mosquitoes. Understanding the genetic factors that regulate gametocyte development is vital for identifying transmission-blocking targets in the malaria parasite life cycle. Toward this end, we conducted a forward genetic study to characterize the development of gametocytes from sexual commitment to mature stage V. We described a new analysis pipeline for the piggyBac transposon-based mutagenesis phenotypic screen to identify genes that influence both early and late gametocyte stages. We classified individual mutants that increased or decreased parasite abundance as the hypoproducer and hyperproducer phenotypes, respectively, revealing distinctive temporal genetic factors early and late in the sexual development cycle. The study identifies that disruption in factors involved in transcription, protein trafficking and DNA repair are associated with decreasing gametocyte production, while modifications in phosphatase activity are linked to hyperproduction of gametocytes. Our study provides an optimized approach on genotype-phenotype evaluation, offering a new resource for understanding potential targets for therapeutic intervention strategies to disrupt transmission.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
References
-
- WHO. World malaria report 2021.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

