Bioluminescent Pseudomonas aeruginosa and Escherichia coli for whole-cell screening of antibacterial and adjuvant compounds
- PMID: 39730767
- PMCID: PMC11681086
- DOI: 10.1038/s41598-024-81926-6
Bioluminescent Pseudomonas aeruginosa and Escherichia coli for whole-cell screening of antibacterial and adjuvant compounds
Abstract
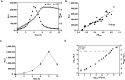
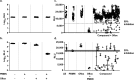
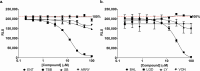
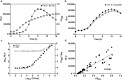
Continued efforts to discover new antibacterial molecules are critical to achieve a robust pre-clinical pipeline for new antibiotics. Screening of compound or natural product extract libraries remains a widespread approach and can benefit from the development of whole cell assays that are robust, simple and versatile, and allow for high throughput testing of antibacterial activity. In this study, we created and validated two bioluminescent reporter strains for high-throughput screening, one in Pseudomonas aeruginosa, and another in a hyperporinated and efflux-deficient Escherichia coli. We show that the bioluminescent strains have a large dynamic range that closely correlates with cell viability and is superior to conventional optical density (OD600) measurements, can detect dose-dependent antibacterial activity and be used for different drug discovery applications. We evaluated the assays' performance characteristics (signal to background ratio, signal window, Z' robust) and demonstrated their potential utility for antibiotic drug discovery in two examples. The P. aeruginosa bioluminescent reporter was used in a pilot screen of 960 repurposed compound libraries to identify adjuvants that potentiate the fluoroquinolone antibiotic ofloxacin. The E. coli bioluminescent reporter was used to test the antibacterial activity of bioactive bacterial supernatants and assist with bioassay-guided fractionation of the crude extracts.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

