Towards a more objective and high-throughput spheroid invasion assay quantification method
- PMID: 39730859
- PMCID: PMC11681046
- DOI: 10.1038/s41598-024-82191-3
Towards a more objective and high-throughput spheroid invasion assay quantification method
Abstract
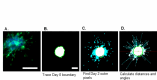
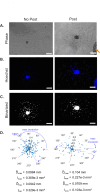
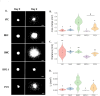
Multicellular spheroids embedded in 3D hydrogels are prominent in vitro models for 3D cell invasion. Yet, quantification methods for spheroid cell invasion that are high-throughput, objective and accessible are still lacking. Variations in spheroid sizes and the shapes of the cells within render it difficult to objectively assess invasion extent. The goal of this work is to develop a high-throughput quantification method of cell invasion into 3D matrices that minimizes sensitivity to initial spheroid size and cell spreading and provides precise integrative directionally-dependent metrics of invasion. By analyzing images of fluorescent cell nuclei, invasion metrics are automatically calculated at the pixel level. The initial spheroid boundary is segmented and automated calculations of the nuclear pixel distances from the initial boundary are used to compute common invasion metrics (i.e., the change in invasion area, mean distance) for the same spheroid at a later timepoint. We also introduce the area moment of inertia as an integrative metric of cell invasion that considers the invasion area as well as the pixel distances from the initial spheroid boundary. Further, we show that principal component analysis can be used to quantify the directional influence of a stimuli to invasion (e.g., due to a chemotactic gradient or contact guidance). To demonstrate the power of the analysis for cell types with different invasive potentials and the utility of this method for a variety of biological applications, the method is used to analyze the invasiveness of five different cell types. In all, implementation of this high-throughput quantification method results in consistent and objective analysis of 3D multicellular spheroid invasion. We provide the analysis code in both MATLAB and Python languages as well as a GUI for ease of use for researchers with a range of computer programming skills and for applications in a variety of biological research areas such as wound healing and cancer metastasis.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Data availability: The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request and are also available at the Harvard Dataverse using the following link: https://doi.org/10.7910/DVN/VQH0BK .
Figures

Update of
-
Towards a More Objective and High-throughput Spheroid Invasion Assay Quantification Method.bioRxiv [Preprint]. 2024 Oct 18:2024.06.27.600893. doi: 10.1101/2024.06.27.600893. bioRxiv. 2024. Update in: Sci Rep. 2024 Dec 28;14(1):31007. doi: 10.1038/s41598-024-82191-3. PMID: 39005385 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

