Dexmedetomidine accelerates photoentrainment and affects sleep structure through the activation of SCNVIP neurons
- PMID: 39730868
- PMCID: PMC11680882
- DOI: 10.1038/s42003-024-07430-9
Dexmedetomidine accelerates photoentrainment and affects sleep structure through the activation of SCNVIP neurons
Abstract
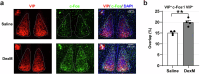
Dexmedetomidine (DexM), a highly selective α2-adrenoceptor agonist, significantly reduces postoperative adverse effects, including sleep and circadian rhythm disruptions. Vasoactive intestinal peptide neurons in the suprachiasmatic nucleus (SCNVIP) regulate the synchronization of circadian rhythms with the external environment in mammals. We investigate the effects of DexM on sleep and circadian rhythms, as well as the underlying mechanisms. Using electrophysiological and chemogenetic methods, along with locomotor activity and electroencephalogram/electromyogram recordings, we found that DexM accelerates the rate of re-entrainment following an 8-hour phase advance in the 12-hour light:12-hour dark cycle, increases the amount of non-rapid eye movement sleep, and decreases the mean duration of rapid eye movement sleep. Chemogenetic inhibition of SCNVIP neurons hinders the acceleration of re-entrainment and the changes in the sleep-wakefulness cycle induced by DexM. Electrophysiological results show that DexM increases the firing rate and the frequency of spontaneous glutamatergic postsynaptic currents while decreasing the frequency of spontaneous GABAergic PSCs in SCNVIP neurons through the α2-adrenergic receptor. Additionally, DexM reduces the frequency of miniature GABAergic PSCs in SCNVIP neurons. In conclusion, these findings suggest that DexM promotes sleep and maintains the coordination of circadian rhythms with the external environment by activating SCNVIP neurons through the α2-adrenoceptor.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

