Therapy response monitoring in blood plasma from esophageal adenocarcinoma patients using cell-free DNA methylation profiling
- PMID: 39730941
- PMCID: PMC11681053
- DOI: 10.1038/s41598-024-82325-7
Therapy response monitoring in blood plasma from esophageal adenocarcinoma patients using cell-free DNA methylation profiling
Abstract
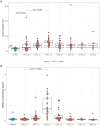
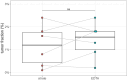
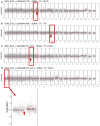
Esophageal adenocarcinoma (EAC) is an aggressive cancer characterized by a high risk of relapse post-surgery. Current follow-up methods (serum carcinoembryonic antigen detection and PET-CT) lack sensitivity and reliability, necessitating a novel approach. Analyzing cell-free DNA (cfDNA) from blood plasma emerges as a promising avenue. This study aims to evaluate the cost-effective and genome-wide cell-free reduced representation bisulfite sequencing (cfRRBS) method combined with computational deconvolution for effective disease monitoring in EAC patients. cfDNA methylation profiling with cfRRBS was performed on 162 blood plasma samples from 33 EAC cancer patients and 28 blood plasma samples from 20 healthy donors. The estimated tumor fraction for EAC patients at the time of diagnosis was significantly different from the healthy donor plasma samples (one-sided Wilcoxon rank-sum test: p-value = 0.032). Tumor fractions above 15% and focal gains/amplifications in MYC (chr8), KRAS (chr12), EGFR (chr7) and NOTCH2 (chr1) were observed in four samples of distinct patients at the time metastatic disease was detected. This study showed feasibility to estimate tumor fractions in blood plasma of EAC patients based on cfDNA methylation using cfRRBS and computational deconvolution. Nevertheless, in this study only cancer patients with evidence of metastatic disease show high tumor fractions and copy number alterations.
Keywords: Blood plasma; DNA methylation; Esophageal adenocarcinoma; Liquid biopsy; cfDNA.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics: Written informed consent was obtained from all cancer patients and healthy donors. Sample collection was approved by the ethics committee of Ghent University Hospital (registration numbers B670201628317, B670201628319 and B670201733701). The research was conducted according to the local legislation and institutional requirements.
Figures

References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

