EDCLoc: a prediction model for mRNA subcellular localization using improved focal loss to address multi-label class imbalance
- PMID: 39731012
- PMCID: PMC11674359
- DOI: 10.1186/s12864-024-11173-6
EDCLoc: a prediction model for mRNA subcellular localization using improved focal loss to address multi-label class imbalance
Abstract
Background: The subcellular localization of mRNA plays a crucial role in gene expression regulation and various cellular processes. However, existing wet lab techniques like RNA-FISH are usually time-consuming, labor-intensive, and limited to specific tissue types. Researchers have developed several computational methods to predict mRNA subcellular localization to address this. These methods face the problem of class imbalance in multi-label classification, causing models to favor majority classes and overlook minority classes during training. Additionally, traditional feature extraction methods have high computational costs, incomplete features, and may lead to the loss of critical information. On the other hand, deep learning methods face challenges related to hardware performance and training time when handling complex sequences. They may suffer from the curse of dimensionality and overfitting problems. Therefore, there is an urgent need for more efficient and accurate prediction models.
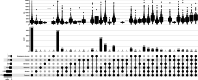
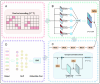
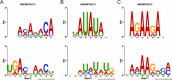
Results: To address these issues, we propose a multi-label classifier, EDCLoc, for predicting mRNA subcellular localization. EDCLoc reduces training pressure through a stepwise pooling strategy and applies grouped convolution blocks of varying sizes at different levels, combined with residual connections, to achieve efficient feature extraction and gradient propagation. The model employs global max pooling at the end to further reduce feature dimensions and highlight key features. To tackle class imbalance, we improved the focal loss function to enhance the model's focus on minority classes. Evaluation results show that EDCLoc outperforms existing methods in most subcellular regions. Additionally, the position weight matrix extracted by multi-scale CNN filters can match known RNA-binding protein motifs, demonstrating EDCLoc's effectiveness in capturing key sequence features.
Conclusions: EDCLoc outperforms existing prediction tools in most subcellular regions and effectively mitigates class imbalance issues in multi-label classification. These advantages make EDCLoc a reliable choice for multi-label mRNA subcellular localization. The dataset and source code used in this study are available at https://github.com/DellCode233/EDCLoc .
Keywords: Class imbalance; Deep learning; Focal loss; MRNA subcellular localization; Multi-label.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
MSlocPRED: deep transfer learning-based identification of multi-label mRNA subcellular localization.Brief Bioinform. 2024 Sep 23;25(6):bbae504. doi: 10.1093/bib/bbae504. Brief Bioinform. 2024. PMID: 39401145 Free PMC article.
-
Deep learning model for protein multi-label subcellular localization and function prediction based on multi-task collaborative training.Brief Bioinform. 2024 Sep 23;25(6):bbae568. doi: 10.1093/bib/bbae568. Brief Bioinform. 2024. PMID: 39489606 Free PMC article.
-
mRNA-CLA: An interpretable deep learning approach for predicting mRNA subcellular localization.Methods. 2024 Jul;227:17-26. doi: 10.1016/j.ymeth.2024.04.018. Epub 2024 May 3. Methods. 2024. PMID: 38705502
-
LncLocFormer: a Transformer-based deep learning model for multi-label lncRNA subcellular localization prediction by using localization-specific attention mechanism.Bioinformatics. 2023 Dec 1;39(12):btad752. doi: 10.1093/bioinformatics/btad752. Bioinformatics. 2023. PMID: 38109668 Free PMC article.
-
Comprehensive review and assessment of computational methods for predicting RNA post-transcriptional modification sites from RNA sequences.Brief Bioinform. 2020 Sep 25;21(5):1676-1696. doi: 10.1093/bib/bbz112. Brief Bioinform. 2020. PMID: 31714956 Review.
References
-
- Buccitelli C, Selbach M. mRNAs, proteins and the emerging principles of gene expression control. Nat Rev Genet. 2020;21:630–44. - PubMed
-
- Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen R-P. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–7. - PubMed
-
- Gonsalvez GB, Urbinati CR, Long RM. RNA localization in yeast: moving towards a mechanism. Biol Cell. 2005;97:75–86. - PubMed
-
- Kugler J-M, Lasko P. Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during Drosophila oogenesis. Fly. 2009;3:15–28. - PubMed
MeSH terms
Substances
Grants and funding
- GJJ2400909,GJJ2402711/the Scientific Research Plan of the Department of Education of Jiangxi Province, China
- GJJ2400909,GJJ2402711/the Scientific Research Plan of the Department of Education of Jiangxi Province, China
- GJJ2400909,GJJ2402711/the Scientific Research Plan of the Department of Education of Jiangxi Province, China
LinkOut - more resources
Full Text Sources
Miscellaneous

