Knockdown of RASD1 improves MASLD progression by inhibiting the PI3K/AKT/mTOR pathway
- PMID: 39731125
- PMCID: PMC11681670
- DOI: 10.1186/s12944-024-02419-z
Knockdown of RASD1 improves MASLD progression by inhibiting the PI3K/AKT/mTOR pathway
Abstract
Background: There is still no reliable therapeutic targets and effective pharmacotherapy for metabolic dysfunction-associated steatotic liver disease (MASLD). RASD1 is short for Ras-related dexamethasone-induced 1, a pivotal factor in various metabolism processes of Human. However, the role of RASD1 remains poorly illustrated in MASLD. Therefore, we designed a study to elucidate how RASD1 could impact on MASLD as well as the mechanisms involved.
Methods: The expression level of RASD1 was validated in MASLD. Lipid metabolism and its underlying mechanism were investigated in hepatocytes and mice with either overexpression or knockdown of RASD1.
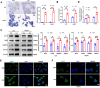
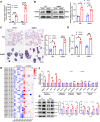
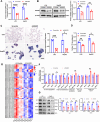
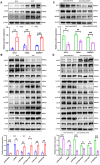
Results: Hepatic RASD1 expression was upregulated in MASLD. Lipid deposition was significantly reduced in RASD1-knockdown hepatocytes and mice, accompanied by a marked downregulation of key genes in the signaling pathway of de novo lipogenesis. Conversely, RASD1 overexpression in hepatocytes had the opposite effect. Mechanistically, RASD1 regulated lipid metabolism in MASLD through the PI3K/AKT/mTOR signaling pathway.
Conclusions: We discovered a novel role of RASD1 in MASLD by regulating lipogenesis via the PI3K/AKT/mTOR pathway, thereby identifying a potential treatment target for MASLD.
Keywords: Lipid metabolism; MASLD; PI3K/AKT/mTOR pathway; Ras-related dexamethasone-induced 1.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The Ethics Committee of the Fifth Affiliated Hospital of Sun Yat-sen University approved this study (approval number: [2020] L173-1). Informed consent was obtained from all patients before surgery. All animal experiments and related protocols were reviewed and approved by the Animal Ethics Committee of the Fifth Affiliated Hospital of Sun Yat-sen University (NO. 00287). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
BRCA1 regulates glucose and lipid metabolism in diabetes mellitus with metabolic dysfunction-associated steatotic liver disease via the PI3K/Akt signaling pathway.PLoS One. 2025 Mar 26;20(3):e0318696. doi: 10.1371/journal.pone.0318696. eCollection 2025. PLoS One. 2025. PMID: 40138287 Free PMC article.
-
MC-LR Aggravates Liver Lipid Metabolism Disorders in Obese Mice Fed a High-Fat Diet via PI3K/AKT/mTOR/SREBP1 Signaling Pathway.Toxins (Basel). 2022 Nov 30;14(12):833. doi: 10.3390/toxins14120833. Toxins (Basel). 2022. PMID: 36548730 Free PMC article.
-
The Regulation of Lipid Deposition by Insulin in Goose Liver Cells Is Mediated by the PI3K-AKT-mTOR Signaling Pathway.PLoS One. 2015 May 6;10(5):e0098759. doi: 10.1371/journal.pone.0098759. eCollection 2015. PLoS One. 2015. PMID: 25945932 Free PMC article.
-
Fanlian Huazhuo Formula: A promising herbal preparation for metabolic liver disease.World J Gastroenterol. 2024 Dec 14;30(46):4964-4968. doi: 10.3748/wjg.v30.i46.4964. World J Gastroenterol. 2024. PMID: 39679304 Free PMC article. Review.
-
Advancing the Metabolic Dysfunction-Associated Steatotic Liver Disease Proteome: A Post-Translational Outlook.Genes (Basel). 2025 Mar 12;16(3):334. doi: 10.3390/genes16030334. Genes (Basel). 2025. PMID: 40149485 Free PMC article. Review.
References
-
- Rinella ME, Lazarus JV, Ratziu V, Ratziu V, Francque SM, Sanyal AJ, et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. J Hepatol. 2023;79:1542–56. - PubMed
-
- Eslam M, Newsome PN, Sarin SK, Anstee QM, Targher G, Romero-Gomez M, et al. A new definition for metabolic associated fatty liver disease: an international expert consensus statement. J Hepatol. 2020;73:202–9. - PubMed
-
- Miao L, Targher G, Byrne CD, Cao YY, Zheng MH. Current status and future trends of the global burden of MASLD. Trends Endocrinol Metab. 2024;29:S1043. -2760(24)00036 – 5. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

