Agrobacterium-mediated transient transformation of Flaveria bidentis leaves: a novel method to examine the evolution of C4 photosynthesis
- PMID: 39731143
- PMCID: PMC11674322
- DOI: 10.1186/s13007-024-01306-z
Agrobacterium-mediated transient transformation of Flaveria bidentis leaves: a novel method to examine the evolution of C4 photosynthesis
Abstract
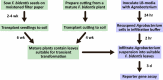
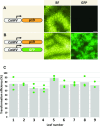
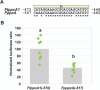
The genus Flaveria has been studied extensively as a model for the evolution of C4 photosynthesis. Thus far, molecular analyses in this genus have been limited due to a dearth of genomic information and the lack of a rapid and efficient transformation protocol. Since their development, Agrobacterium-mediated transient transformation protocols have been instrumental in understanding many biological processes in a range of plant species. However, this technique has not been applied to the genus Flaveria. Here, an efficient protocol for the Agrobacterium-mediated transient transformation of the leaves of the C4 species Flaveria bidentis is presented. This technique has the distinct advantages of rapid turnaround, the ability to co-transform with multiple constructs, and the capacity to assay coding and non-coding regions of Flaveria genomes in a homologous context. To illustrate the utility of this protocol, the quantitative transcriptional regulation of phosphoenolpyruvate carboxylase, the primary carboxylase of C4 plants, was investigated. A 24 bp region in the ppcA1 proximal promoter was found to elicit high levels of reporter gene expression. The Agrobacterium-mediated transient transformation of F. bidentis leaves will accelerate the understanding of the biology and evolution of C4 photosynthesis in the genus Flaveria as well as in other C4 lineages.
Keywords: Flaveria; C4 photosynthesis; Evolution of C4 photosynthesis; Phosphoenolpyruvate carboxylase; Transient transformation.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





References
-
- Osmond CB, Winter K, Ziegler H. Functional significance of different pathways of CO2 fixation in photosynthesis. In: Lange OL, Nobel PS, Osmond CB, Ziegler H, editors. Physiological plant ecology II. Encyclopedia of plant physiology. Berlin: Springer; 1982. p. 479–547.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

