Metagenome-validated combined amplicon sequencing and text mining-based annotations for simultaneous profiling of bacteria and fungi: vaginal microbiota and mycobiota in healthy women
- PMID: 39731160
- PMCID: PMC11681650
- DOI: 10.1186/s40168-024-01993-9
Metagenome-validated combined amplicon sequencing and text mining-based annotations for simultaneous profiling of bacteria and fungi: vaginal microbiota and mycobiota in healthy women
Abstract
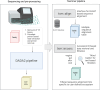
Background: Amplicon sequencing of kingdom-specific tags such as 16S rRNA gene for bacteria and internal transcribed spacer (ITS) region for fungi are widely used for investigating microbial communities. So far most human studies have focused on bacteria while studies on host-associated fungi in health and disease have only recently started to accumulate. To enable cost-effective parallel analysis of bacterial and fungal communities in human and environmental samples, we developed a method where 16S rRNA gene and ITS1 amplicons were pooled together for a single Illumina MiSeq or HiSeq run and analysed after primer-based segregation. Taxonomic assignments were performed with Blast in combination with an iterative text-extraction-based filtration approach, which uses extensive literature records from public databases to select the most probable hits that were further validated by shotgun metagenomic sequencing.
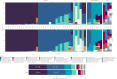
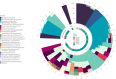
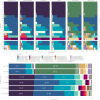
Results: Using 50 vaginal samples, we show that the combined run provides comparable results on bacterial composition and diversity to conventional 16S rRNA gene amplicon sequencing. The text-extraction-based taxonomic assignment-guided tool provided ecosystem-specific bacterial annotations that were confirmed by shotgun metagenomic sequencing (VIRGO, MetaPhlAn, Kraken2). Fungi were identified in 39/50 samples with ITS sequencing while in the metagenome data fungi largely remained undetected due to their low abundance and database issues. Co-abundance analysis of bacteria and fungi did not show strong between-kingdom correlations within the vaginal ecosystem of healthy women.
Conclusion: Combined amplicon sequencing for bacteria and fungi provides a simple and cost-effective method for simultaneous analysis of microbiota and mycobiota within the same samples. Conventional metagenomic sequencing does not provide sufficient fungal genome coverage for their reliable detection in vaginal samples. Text extraction-based annotation tool facilitates ecosystem-specific characterization and interpretation of microbial communities by coupling sequence homology to microbe metadata readily available through public databases. Video Abstract.
Keywords: 16S rRNA gene; ITS; Metagenomics; Microbiota; Mycobiota; Parallel sequencing; Text mining; Vagina.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was approved by the ethical committee of The Hospital District of Helsinki and Uusimaa and Helsinki region hospital district (21/13/03/03/2014) and performed in accordance with the principles of the Helsinki Declaration. All participants signed an informed consent. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

