Structural Immunology of SARS-CoV-2
- PMID: 39731211
- PMCID: PMC11727448
- DOI: 10.1111/imr.13431
Structural Immunology of SARS-CoV-2
Abstract
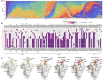
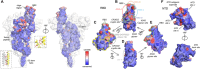
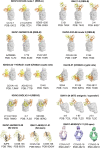
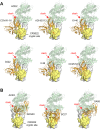
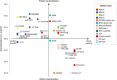
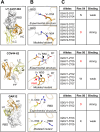
The SARS-CoV-2 spike (S) protein has undergone significant evolution, enhancing both receptor binding and immune evasion. In this review, we summarize ongoing efforts to develop antibodies targeting various epitopes of the S protein, focusing on their neutralization potency, breadth, and escape mechanisms. Antibodies targeting the receptor-binding site (RBS) typically exhibit high neutralizing potency but are frequently evaded by mutations in SARS-CoV-2 variants. In contrast, antibodies targeting conserved regions, such as the S2 stem helix and fusion peptide, exhibit broader reactivity but generally lower neutralization potency. However, several broadly neutralizing antibodies have demonstrated exceptional efficacy against emerging variants, including the latest omicron subvariants, underscoring the potential of targeting vulnerable sites such as RBS-A and RBS-D/CR3022. We also highlight public classes of antibodies targeting different sites on the S protein. The vulnerable sites targeted by public antibodies present opportunities for germline-targeting vaccine strategies. Overall, developing escape-resistant, potent antibodies and broadly effective vaccines remains crucial for combating future variants. This review emphasizes the importance of identifying key epitopes and utilizing antibody affinity maturation to inform future therapeutic and vaccine design.
Keywords: COVID‐19; SARS‐CoV‐2; neutralizing antibody; spike; vaccine; viral evolution.
© 2024 The Author(s). Immunological Reviews published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- WHO , “WHO COVID‐19 Dashboard,” https://data.who.int/dashboards/covid19/cases.
-
- Letunic I. and Bork P., “Interactive Tree of Life (iTOL): An Online Tool for Phylogenetic Tree Display and Annotation,” Bioinformatics 23, no. 1 (2007): 127–128. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

