Coupling of response biomarkers between tumor and peripheral blood in patients undergoing chemoimmunotherapy
- PMID: 39731918
- PMCID: PMC11866441
- DOI: 10.1016/j.xcrm.2024.101882
Coupling of response biomarkers between tumor and peripheral blood in patients undergoing chemoimmunotherapy
Abstract
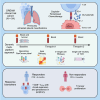
Platinum-based chemotherapy in combination with anti-PD-L1 antibodies has shown promising results in mesothelioma. However, the immunological mechanisms underlying its efficacy are not well understood and there are no predictive biomarkers to guide treatment decisions. Here, we combine time course RNA sequencing (RNA-seq) of peripheral blood mononuclear cells with pre-treatment tumor transcriptome data from the single-arm, phase 2 DREAM trial (N = 54). Single-cell RNA-seq and T cell receptor sequencing (TCR-seq) reveal that CD8+ T effector memory (TEM) cells with stem-like properties are more abundant in peripheral blood of responders and that this population expands upon treatment. These peripheral blood changes are linked to the transcriptional state of the tumor microenvironment. Combining information from both compartments, rather than individually, is most predictive of response. Our study highlights complex interactions between the tumor and immune cells in peripheral blood during objective tumor responses to chemoimmunotherapy. This trial is registered with the Australian New Zealand Clinical Trials Registry, number ACTRN12616001170415.
Keywords: CD8(+) T effector memory cells; ICT; PD-L1; biomarkers; cancer; immune checkpoint therapy; mesothelioma; peripheral blood; platinum chemotherapy; response; single-cell RNA sequencing.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests W.J.L. declares consultancy for Douglas Pharmaceuticals and research funding from Douglas Pharmaceuticals, AstraZeneca, and ENA therapeutics. W.J.L. is a director of the Cancer Research Trust, which has funded parts of this work, but is not involved in any funding decisions, which are based on independent external peer review. W.J.L. is a founder and director of Setonix Pharmaceuticals, which is not related to the presented work. W.J.L. is an inventor on patent applications unrelated to the presented work. A.K.N. declares consultancy for Douglas Pharmaceuticals and Bristol-Myers Squibb, institutional research funding from AstraZeneca, and consultancy (unrelated to this work) from AstraZeneca.
Figures




References
-
- Vogelzang N.J., Rusthoven J.J., Symanowski J., Denham C., Kaukel E., Ruffie P., Gatzemeier U., Boyer M., Emri S., Manegold C., et al. Phase III study of pemetrexed in combination with cisplatin versus cisplatin alone in patients with malignant pleural mesothelioma. J. Clin. Oncol. 2003;21:2636–2644. doi: 10.1200/JCO.2003.11.136. - DOI - PubMed
-
- Baas P., Scherpereel A., Nowak A.K., Fujimoto N., Peters S., Tsao A.S., Mansfield A.S., Popat S., Jahan T., Antonia S., et al. First-line nivolumab plus ipilimumab in unresectable malignant pleural mesothelioma (CheckMate 743): a multicentre, randomised, open-label, phase 3 trial. Lancet. 2021;397:375–386. doi: 10.1016/S0140-6736(20)32714-8. - DOI - PubMed
-
- Nowak A.K., Lesterhuis W.J., Kok P.-S., Brown C., Hughes B.G., Karikios D.J., John T., Kao S.C.-H., Leslie C., Cook A.M., et al. Durvalumab with first-line chemotherapy in previously untreated malignant pleural mesothelioma (DREAM): a multicentre, single-arm, phase 2 trial with a safety run-in. Lancet Oncol. 2020;21:1213–1223. doi: 10.1016/S1470-2045(20)30462-9. - DOI - PubMed
-
- Kok P.S., Forde P.M., Hughes B., Sun Z., Brown C., Ramalingam S., Cook A., Lesterhuis W.J., Yip S., O’Byrne K., et al. Protocol of DREAM3R: DuRvalumab with chEmotherapy as first-line treAtment in advanced pleural Mesothelioma—a phase 3 randomised trial. BMJ Open. 2022;12 doi: 10.1136/bmjopen-2021-057663. - DOI - PMC - PubMed
-
- Piccirillo M.C., Chu Q., Bradbury P., Tu W., Coschi C.H., Grosso F., Florescu M., Mencoboni M., Goffin J.R., Pagano M., et al. Brief Report: Canadian Cancer Trials Group IND.227: A Phase 2 Randomized Study of Pembrolizumab in Patients With Advanced Malignant Pleural Mesothelioma (NCT02784171) J. Thorac. Oncol. 2023;18:813–819. doi: 10.1016/j.jtho.2023.02.003. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

