Computational Pathology Detection of Hypoxia-Induced Morphologic Changes in Breast Cancer
- PMID: 39732389
- PMCID: PMC12179535
- DOI: 10.1016/j.ajpath.2024.10.023
Computational Pathology Detection of Hypoxia-Induced Morphologic Changes in Breast Cancer
Abstract
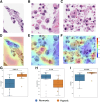
Understanding the tumor hypoxic microenvironment is crucial for grasping tumor biology, clinical progression, and treatment responses. This study presents a novel application of artificial intelligence in computational histopathology to evaluate hypoxia in breast cancer. Weakly supervised deep learning models can accurately detect morphologic changes associated with hypoxia in routine hematoxylin and eosin (H&E)-stained whole slide images (WSIs). The HypOxNet model was trained on H&E-stained WSIs from breast cancer primary sites (n = 1016) at ×40 magnification using data from The Cancer Genome Atlas. Hypoxia Buffa signature was used to measure hypoxia scores, which ranged from -43 to 47, and stratified the samples into hypoxic and normoxic based on these scores. This stratification represented the weak labels associated with each WSI. HypOxNet achieved an average area under the curve of 0.82 on test sets, identifying significant differences in cell morphology between hypoxic and normoxic tissue regions. Importantly, once trained, the HypOxNet model required only the readily available H&E-stained slides, making it especially valuable in low-resource settings where additional gene expression assays are not available. These artificial intelligence-based hypoxia detection models can potentially be extended to other tumor types and seamlessly integrated into pathology workflows, offering a fast, cost-effective alternative to molecular testing.
Copyright © 2025 American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Disclosure Statement None declared.
Figures




References
-
- Tutzauer J., Sjöström M., Holmberg E., Karlsson P., Killander F., Leeb-Lundberg L.M.F., Malmström P., Niméus E., Fernö M., Jögi A. Breast cancer hypoxia in relation to prognosis and benefit from radiotherapy after breast-conserving surgery in a large, randomised trial with long-term follow-up. Br J Cancer. 2022;126:1145–1156. - PMC - PubMed
-
- Bhandari V., Hoey C., Liu L.Y., Lalonde E., Ray J., Livingstone J., Lesurf R., Shiah Y.-J., Vujcic T., Huang X., Espiritu S.M.G., Heisler L.E., Yousif F., Huang V., Yamaguchi T.N., Yao C.Q., Sabelnykova V.Y., Fraser M., Chua M.L.K., van der Kwast T., Liu S.K., Boutros P.C., Bristow R.G. Molecular landmarks of tumor hypoxia across cancer types. Nat Genet. 2019;51:308–318. - PubMed
-
- Vaupel P., Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26:225–239. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

