Antimicrobial resistance, virulence gene profiling, and genetic diversity of multidrug-resistant Pseudomonas aeruginosa isolates in Mazandaran, Iran
- PMID: 39732629
- PMCID: PMC11681713
- DOI: 10.1186/s12866-024-03707-5
Antimicrobial resistance, virulence gene profiling, and genetic diversity of multidrug-resistant Pseudomonas aeruginosa isolates in Mazandaran, Iran
Erratum in
-
Correction: Antimicrobial resistance, virulence gene profiling, and genetic diversity of multidrug-resistant Pseudomonas aeruginosa isolates in Mazandaran, Iran.BMC Microbiol. 2025 Feb 1;25(1):58. doi: 10.1186/s12866-025-03787-x. BMC Microbiol. 2025. PMID: 39891045 Free PMC article. No abstract available.
Abstract
Background: Pseudomonas aeruginosa is a major cause of healthcare-associated infections (HAIs), particularly in immunocompromised patients, leading to high morbidity and mortality rates. This study aimed to investigate the antimicrobial resistance patterns, virulence gene profiles, and genetic diversity among P. aeruginosa isolates from hospitalized patients in Mazandaran, Iran.
Methods: From September 2021 to April 2022, 82 non-duplicate P. aeruginosa isolates were collected from diverse clinical sources. Identification was confirmed using API 20 NE (bioMérieux, Marcy l'Etoile, France). Antimicrobial susceptibility testing was conducted using the Kirby-Bauer disk diffusion method according to CLSI guidelines to assess resistance to a range of antibiotics. The virulence profile (exoT, exoY, exoU, toxA, plcH, plcN, algD, aprA, lasB and exoS) of each P. aeruginosa isolate was determined by PCR. The genetic diversity among the strains was evaluated using the random amplification of polymorphic DNA (RAPD) technique. Clustering was based on a Dice similarity coefficient of ≥ 85%.
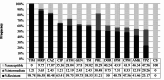
Results: Of the 82 total strains, P. aeruginosa exhibited the highest and lowest resistance toward ticarcillin-clavulanate (98.78%) and colistin (0%), respectively. Moreover, 100% of the P. aeruginosa isolates were MDR. The following prevalence of virulence factor genes was observed: aprA, lasB, algD, toxA, plcH, exoY, and exoT in 100% of isolates. The plcN, exoS, and exoU were identified 98.78%, 67.07%, and 45.12%, respectively. The RAPD patterns obtained with primers 272 and 208 had respectively 2-19 and 6-17 bands. According to the Dice similarity coefficient of higher than 85%, 56 and 39 clusters were recognized.
Conclusion: The high rate of multidrug resistance combined with the widespread presence of virulence genes in P. aeruginosa isolates highlights the potential for increased infection severity, morbidity, and mortality in hospitalized patients. The substantial genetic diversity observed among isolates suggests that P. aeruginosa in this region may rapidly evolve, necessitating ongoing surveillance and more targeted antimicrobial strategies.
Clinical trial number: Not applicable.
Keywords: P. aeruginosa; Antibiotic resistance; Multidrug-resistant; RAPD; Virulence gene.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was performed in agreement with the Declaration of Helsinki. This study was also approved by the Iran National Committee for Ethics in Biomedical Research with the national ethical code (ref number: IR.MAZUMS.REC.1399.901) and informed consent was obtained from all participants and/or their guardian. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- Strateva T, Yordanov D. Pseudomonas aeruginosa–a phenomenon of bacterial resistance. J Med Microbiol. 2009;58(9):1133–48. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

