THBS2 + cancer-associated fibroblasts promote EMT leading to oxaliplatin resistance via COL8A1-mediated PI3K/AKT activation in colorectal cancer
- PMID: 39732719
- PMCID: PMC11681647
- DOI: 10.1186/s12943-024-02180-y
THBS2 + cancer-associated fibroblasts promote EMT leading to oxaliplatin resistance via COL8A1-mediated PI3K/AKT activation in colorectal cancer
Erratum in
-
Correction: THBS2 + cancer-associated fibroblasts promote EMT leading to oxaliplatin resistance via COL8A1-mediated PI3K/AKT activation in colorectal cancer.Mol Cancer. 2025 Jun 26;24(1):184. doi: 10.1186/s12943-025-02388-6. Mol Cancer. 2025. PMID: 40571948 Free PMC article. No abstract available.
Abstract
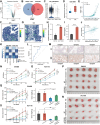
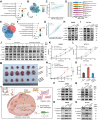
Cancer-associated fibroblasts (CAFs) exert multiple tumor-promoting functions and are key contributors to drug resistance. The mechanisms by which specific subsets of CAFs facilitate oxaliplatin resistance in colorectal cancer (CRC) have not been fully explored. This study found that THBS2 is positively associated with CAF activation, epithelial-mesenchymal transition (EMT), and chemoresistance at the pan-cancer level. Together with single-cell RNA sequencing and spatial transcriptomics analyses, we identified THBS2 specifically derived from subsets of CAFs, termed THBS2 + CAFs, which could promote oxaliplatin resistance by interacting with malignant cells via the collagen pathway in CRC. Mechanistically, COL8A1 specifically secreted from THBS2 + CAFs directly interacts with the ITGB1 receptor on resistant malignant cells, activating the PI3K-AKT signaling pathway and promoting EMT, ultimately leading to oxaliplatin resistance in CRC. Moreover, elevated COL8A1 promotes EMT and contributes to CRC oxaliplatin resistance, which can be mitigated by ITGB1 knockdown or AKT inhibitor. Collectively, these results highlight the crucial role of THBS2 + CAFs in promoting oxaliplatin resistance of CRC by activating EMT and provide a rationale for a novel strategy to overcome oxaliplatin resistance in CRC.
Keywords: COL8A1; Cancer-associated fibroblasts; Colorectal cancer; EMT; Oxaliplatin resistance.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Consent for publication: All authors have consented to submit this article for publication. Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

