Bioinformatics identifies key genes and potential therapeutic targets in the pathological mechanism of oxidative stress in Randall's plaque
- PMID: 39732836
- PMCID: PMC11682209
- DOI: 10.1038/s41598-024-82849-y
Bioinformatics identifies key genes and potential therapeutic targets in the pathological mechanism of oxidative stress in Randall's plaque
Abstract
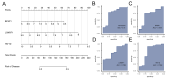
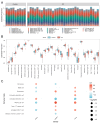
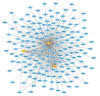
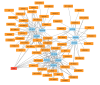
Randall's plaque (RP) is recognized as a precursor lesion for kidney stones, with its formation and progression potentially linked to oxidative stress. Previous studies have provided limited insights into the underlying mechanisms of RP, failing to fully elucidate its molecular pathways. To investigate the relationship between oxidative stress and RP, we employed bioinformatics approaches to identify key genes, predict associated pathways and drug molecules, analyze variations in immune cell populations, and construct diagnostic models. We initially identified three differentially expressed genes related to oxidative stress: BFSP1, LONF1, and TAF1D. These genes and their co-expressed counterparts are enriched in pathways related to oxidative phosphorylation, cellular adhesion processes, steroid hormone biosynthesis, and autophagy. Furthermore, we observed significant differences in two types of immune cells across the study groups. Ultimately, predictions from drug molecular docking suggest that BFSP1 may serve as a promising therapeutic target for RP. We propose that the formation of RP mediated by oxidative stress could be associated with BFSP1, LONF1, TAF1D along with CD56dim natural killer cells and memory B cells. Thus far, BFSP1 emerges as a pivotal therapeutic target for RP development. These findings offer new perspectives on the mechanisms underlying the pathogenesis of RP.
Keywords: BFSP1; Bioinformatic analysis; LONRF1; Oxidative stress; Randall's plaque; TAF1D.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
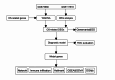
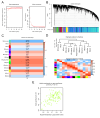

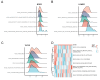
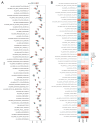
References
MeSH terms
LinkOut - more resources
Full Text Sources

