Integrating immune multi-omics and machine learning to improve prognosis, immune landscape, and sensitivity to first- and second-line treatments for head and neck squamous cell carcinoma
- PMID: 39732954
- PMCID: PMC11682253
- DOI: 10.1038/s41598-024-83184-y
Integrating immune multi-omics and machine learning to improve prognosis, immune landscape, and sensitivity to first- and second-line treatments for head and neck squamous cell carcinoma
Abstract
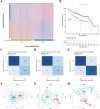
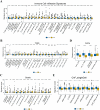
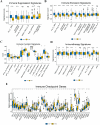
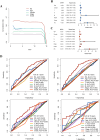
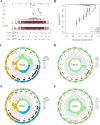
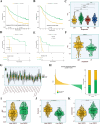
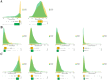
In recent years, immune checkpoint inhibitors (ICIs) has emerged as a fundamental component of the standard treatment regimen for patients with head and neck squamous cell carcinoma (HNSCC). However, accurately predicting the treatment effectiveness of ICIs for patients at the same TNM stage remains a challenge. In this study, we first combined multi-omics data (mRNA, lncRNA, miRNA, DNA methylation, and somatic mutations) and 10 clustering algorithms, successfully identifying two distinct cancer subtypes (CSs) (CS1 and CS2). Subsequently, immune-regulated genes (IRGs) and machine learning algorithms were utilized to construct a consensus machine learning-driven prediction immunotherapy signature (CMPIS). Further, the prognostic model was validated and compared across multiple datasets, including clinical characteristics, external datasets, and previously published models. Ultimately, the response of different CMPIS patients to immunotherapy, targeted therapy, radiotherapy and chemotherapy was also explored. First, Two distinct molecular subtypes were successfully identified by integrating immunomics data with machine learning techniques, and it was discovered that the CS1 subtype tended to be classified as "cold tumors" or "immunosuppressive tumors", whereas the CS2 subtype was more likely to represent "hot tumors" or "immune-activated tumors". Second, 303 different algorithms were employed to construct prognostic models and the average C-index value for each model was calculated across various cohorts. Ultimately, the StepCox [forward] + Ridge algorithm, which had the highest average C-index value of 0.666, was selected and this algorithm was used to construct the CMPIS predictive model comprising 16 key genes. Third, this predictive model was compared with patients' clinical features, such as age, gender, TNM stage, and grade stage. The findings indicated that this prognostic model exhibited the best performance in terms of C-index and AUC values. Additionally, it was compared with previously published models and it was found that the C-index of CMPIS ranked in the top 5 among 94 models across the TCGA, GSE27020, GSE41613, GSE42743, GSE65858, and META datasets. Lastly, the study revealed that patients with lower CMPIS were more sensitive to immunotherapy and chemotherapy, while those with higher CMPIS were more responsive to radiation therapy and EGFR-targeted treatments. In summary, our study identified two CSs (CS1 and CS2) of HNSCC using multi-omics data and predicted patient prognosis and treatment response by constructing the CMPIS model with IRGs and 303 machine learning algorithms, which underscores the importance of immunotherapy biomarkers in providing more targeted, precise, and personalized immunotherapy plans for HNSCC patients, significantly contributing to the optimization of clinical treatment outcomes.
Keywords: Consensus machine learning-driven prediction immunotherapy signature; Head and neck squamous cell carcinoma; Immune checkpoint inhibitors; Machine learning; Multi-omics; Prognosis.
© 2024. The Author(s).
Conflict of interest statement
Declaration. Competing interests: The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
- 2023M730378/The China Postdoctoral Science Foundation
- 2023M730378/The China Postdoctoral Science Foundation
- BSH2023025/Xinglin Scholar" Scientific Research Promotion Plan of Chengdu University of TCM
- BSH2023025/Xinglin Scholar" Scientific Research Promotion Plan of Chengdu University of TCM
- 2024NSFC0722/Sichuan Province Science and Technology Support Program
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

