Infectious agent release and Pacific salmon exposure at Atlantic salmon farms revealed by environmental DNA
- PMID: 39732981
- PMCID: PMC11682043
- DOI: 10.1038/s41598-024-83250-5
Infectious agent release and Pacific salmon exposure at Atlantic salmon farms revealed by environmental DNA
Abstract
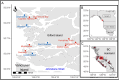
The potential risk posed by infectious agents (IAs) associated with netpen aquaculture to wild fishes is determined based on the "release" of IAs from netpens into the environment, the "exposure" of the wild fish to those released agents, and the "consequence" for wild fish experiencing infection by those agents. Information available to characterize these three factors is often lacking, and the occurrence of transmission from aquaculture to wild fish as well as potential consequences of such transmission are difficult to observe. In this study, we utilized environmental DNA (eDNA) to characterize the release of dozens of IAs from, and exposure of Pacific salmon to, Atlantic salmon aquaculture. We combined these factors with the consequence of infection, as determined by the literature, to identify IAs that may pose a risk to wild salmon exposed to aquaculture in British Columbia, Canada. Over an 18-month period, eDNA samples were collected from seven active and four inactive netpen aquaculture sites in the Broughton Archipelago, BC. A meta-analytical mean across 22 IAs showed that the odds of IA detection at active sites was 4.3 (95% confidence interval = 2.3:8.1) times higher than at inactive sites, with 11 IAs in particular demonstrating a pattern consistent with elevated release. Oncorhynchus tshawytscha was the only Pacific salmon species presenting eDNA detections more likely to occur around and within active netpens relative to inactive sites. After considering the evidence of negative consequences of infection (from previous literature) in tandem with release model results, we determined that Tenacibaculum maritimum, Tenacibaculum finnmarkense, Ichthyobodo spp., and Piscine orthoreovirus are potential risks to Pacific salmon exposed to marine netpen aquaculture. These IAs, and others demonstrating patterns consistent with release but with insufficient prior research to evaluate the consequences of infection, require further studies that identify the factors influencing the intensity of release, the spatial extent of release around netpens, and the prevalence of infection in wild fish within known distances from netpens.
© 2024. Crown.
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures






References
-
- Oidtmann, B., Dixon, P., Way, K., Joiner, C. & Bayley, A. E. Risk of waterborne virus spread-review of survival of relevant fish and crustacean viruses in the aquatic environment and implications for control measures. Rev. Aquac.10, 641–669 (2018). - DOI
-
- Lyngstad, T. M., Høgåsen, H. R., Jansen, M. D. & Nilsen, A. Risk of disease transfer with wellboats in Norway–technical report. Veterinærinstituttets Oslo Rapportserie 15–2015 (2015).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

