Imaging phenotype reveals that disulfirams induce protein insolubility in the mitochondrial matrix
- PMID: 39733149
- PMCID: PMC11682119
- DOI: 10.1038/s41598-024-82939-x
Imaging phenotype reveals that disulfirams induce protein insolubility in the mitochondrial matrix
Abstract
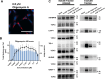
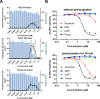
The cell painting assay is useful for understanding cellular phenotypic changes and drug effects. To identify other aspects of well-known chemicals, we screened 258 compounds with the cell painting assay and focused on a mitochondrial punctate phenotype seen with disulfiram. To elucidate the reason for this punctate phenotype, we looked for clues by examining staining steps and gene knockdown as well as examining protein solubility and comparing cell lines. From these results, we found that the punctate phenotype was caused by protein insolubility in the mitochondrial matrix. Interestingly, the punctate phenotype of disulfiram was sensitive to the relative expression of LonP1, a protease in the mitochondrial matrix that regulates proteostasis, suggesting that the punctate phenotype manifests when the protein quality control capacity in the mitochondrial matrix is exceeded. Moreover, we discovered that disulfiram and its derivatives, which have all been reported to increase acetaldehyde in the blood after the in vivo intake of alcohol, induced a punctate phenotype as well. The investigated punctate phenotype not only provides a novel clue for elucidating the common mechanism of action among disulfiram derivatives but is also a novel example of chemical perturbation of proteostasis in the mitochondrial matrix.
Keywords: Cell painting assay; Disulfiram; LonP1; Oligomycin A; Protein insolubility; Proteostasis.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

