In Silico and In Vitro Study of mRNA Biomarkers for Glioblastoma Multiforme Resistance to Temozolomide (TMZ): The Association with Stemness
- PMID: 39733437
- PMCID: PMC12008334
- DOI: 10.31557/APJCP.2024.25.12.4435
In Silico and In Vitro Study of mRNA Biomarkers for Glioblastoma Multiforme Resistance to Temozolomide (TMZ): The Association with Stemness
Abstract
Background: Glioblastoma multiforme (GBM) is an aggressive brain tumor that primarily affects adults. The Stupp Protocol, which includes surgical resection, chemoradiation, and monotherapy with temozolomide (TMZ), is the standard treatment regimen for GBM. However, repeated use of TMZ leads to resistance in GBM cells, resulting in a poor prognosis for patients. This resistance is driven by several intrinsic factors. This study aims to identify potential biomarkers of resistance associated with stemness.
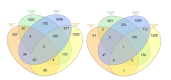
Methods: We utilized datasets from GEO, performed Venn diagram intersection analysis, conducted GO enrichment analysis using DAVID and ENRICHR, carried out pathway enrichment analysis with KEGG and REACTOME, and executed survival analysis with GEPIA. Additionally, we compared mRNA expression using the Human Protein Atlas and validated our findings with qRT-PCR.
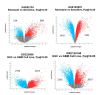
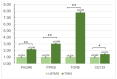
Results: We identified that PAQR6 and ITPKB mRNA expression was consistently higher in TMZ-resistant T98G cells, but TGFBI mRNA expression was found to be significantly higher in TMZ-resistant T98G cells compared to U87MG cells. In addition, a significantly higher CD133 mRNA expression as a stemness marker was found in T98G cells compared to U87MG cells. It is hoped that the acquired disease-related resistance biomarker candidates will be able to be used at the clinical level in terms of non-invasive early detection in GBM patients. However, additional research is required to validate the findings of this preliminary biomarker discovery study.
Keywords: DEG; Enrichment; Glioblastoma; Resistance; stemness.
Conflict of interest statement
The authors confirm that they have no conflicts of interest to disclose.
Figures


References
-
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N Engl J Med. 2005;352(10):987–96. - PubMed
-
- Wang Y, Zhang J, Li W, Jiang T, Qi S, Chen Z, et al. Guideline conformity to the Stupp regimen in patients with newly diagnosed glioblastoma multiforme in China. Futur Oncol. 2021;17(33):4571–82. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

