Exploration of the mechanism of 5-Methylcytosine promoting the progression of hepatocellular carcinoma
- PMID: 39733743
- PMCID: PMC11743813
- DOI: 10.1016/j.tranon.2024.102257
Exploration of the mechanism of 5-Methylcytosine promoting the progression of hepatocellular carcinoma
Abstract
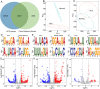
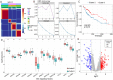
5-Methylcytosine (m5C) is a ubiquitous RNA modification that is closely related to various cellular functions. However, no studies have comprehensively demonstrated the role of m5C in hepatocellular carcinoma (HCC) progression. In this study, six pairs of HCC and adjacent tissue samples were subjected to methylated RNA immunoprecipitation sequencing to identify precise m5C loci. Non-negative matrix factorization (NMF) was used to identify HCC subtypes in TCGA-LIHC cohort. Immune, metabolic, and tumor-related pathways in HCC subtypes with differences in methylation status were analyzed and a prognostic model based on m5C-related genes was constructed. Finally, using RIP and molecular interaction analysis, we demonstrated that YBX1 binds to TPM3 in an m5C dependent manner and regulates HCC progression. Widespread m5C sites were identified and found to be differentially distributed in HCC compared with adjacent tissues. Metabolic processes were inhibited in hypermethylated HCC, whereas immune checkpoint and multiple classical tumor pathways were significantly upregulated. More importantly, we have identified an m5C dependent regulatory axis. The m5C reader YBX1 binds to TPM3 in an M5C dependent manner and promotes the progression of hepatocellular carcinoma. These results provide new evidence for further understanding the comprehensive role of m5C in HCC and the regulatory mechanism of m5C.
Keywords: 5-methylcytosine; HCC; Immunity; Metabolism; Tumor-related pathways; YBX1.
Copyright © 2024. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






References
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. - PubMed
-
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

