Identification of virus-rich intermediate cells as crucial players in SARS-CoV-2 infection and differentiation dynamics of human airway epithelium
- PMID: 39735182
- PMCID: PMC11681626
- DOI: 10.3389/fmicb.2024.1507852
Identification of virus-rich intermediate cells as crucial players in SARS-CoV-2 infection and differentiation dynamics of human airway epithelium
Abstract
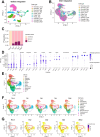
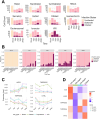
Understanding the early interactions between severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and human airway epithelial cells is essential for unraveling viral replication and spread mechanisms. In this study, we investigated the early dynamics of airway epithelial cells during SARS-CoV-2 infection using well-differentiated human nasal and tracheal epithelial cell cultures by incorporating three publicly available single-cell RNA sequencing datasets. We identified a previously uncharacterized cell population, termed virus-rich intermediate (VRI) cells, representing an intermediate differentiation stage between basal and ciliated cells. These VRI cells exhibited high viral loads at all infection time points, strong interferon and inflammatory responses, increased mRNA expression of microvilli-related genes (PAK1, PAK4, VIL1), and suppression of apoptosis markers (BAX, CASP3) alongside increased anti-apoptotic gene expression (BCL2). Cell-cell interaction analysis revealed that VRI cells send signals to basal cells via receptor-ligand pathways such as EPHA and VEGF, likely promoting basal cell differentiation and proliferation through MAPK signaling. These findings suggest that SARS-CoV-2 utilizes VRI cells as a primary site for replication and spread, leveraging these cells' unique differentiation state to evade host cell death and facilitate viral propagation. This study provides insights into the early cellular responses to SARS-CoV-2 infection and highlights potential therapeutic targets to limit viral spread.
Keywords: SARS-CoV-2; air-liquid interface culture; airway epithelial cell; single-cell RNA sequencing; virus-rich intermediate.
Copyright © 2024 Kim and Lee.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

