Reviewing advancement in Mycoplasma pneumoniae P30 adhesin protein provides insights for future diagnosis and treatment
- PMID: 39735188
- PMCID: PMC11671514
- DOI: 10.3389/fmicb.2024.1515291
Reviewing advancement in Mycoplasma pneumoniae P30 adhesin protein provides insights for future diagnosis and treatment
Abstract
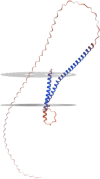
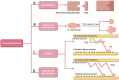
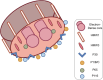
Mycoplasma pneumoniae is a major pathogen that causes upper and lower respiratory tract infections in children, adolescents, and elderly individuals and can lead to pneumonia, intrapulmonary and extrapulmonary complications, and respiratory sequelae. M. pneumoniae must adhere to respiratory epithelial cells of a host for infection. The P1 and P30 proteins, as two adhesin proteins of M. pneumoniae, have attracted extensive attention from many researchers. In this paper, we present the latest research progress on the P30 protein in terms of structure and mutation typing, physiological function, clinical serological diagnosis and vaccine development in a literature review. This study deepens our knowledge on the pathogenesis of M. pneumoniae and is useful for diagnosing and preventing M. pneumoniae infection.
Keywords: Mycoplasma pneumoniae; P30 protein; adhesion; diagnosis; vaccine.
Copyright © 2024 Zuo, Zhang and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Baseman J. B. (1993). “The Cytadhesins of Mycoplasma pneumoniae and M. genitalium” in Mycoplasma Cell Membranes. eds. Rottem S., Kahane I. (Boston, MA: Springer, US; ), 243–259. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

