Rapid, sensitive, and visual detection of mandarin fish ranavirus and infectious spleen and kidney necrosis virus using an RPA-CRISPR/Cas12a system
- PMID: 39735189
- PMCID: PMC11671746
- DOI: 10.3389/fmicb.2024.1495777
Rapid, sensitive, and visual detection of mandarin fish ranavirus and infectious spleen and kidney necrosis virus using an RPA-CRISPR/Cas12a system
Abstract
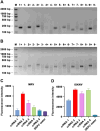
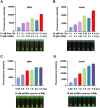
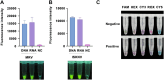
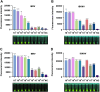
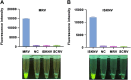
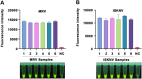
Iridoviruses are large cytoplasmic icosahedral viruses that contain dsDNA. Among them, mandarin fish ranavirus (MRV) and infectious spleen and kidney necrosis virus (ISKNV) are particularly notable due to their high contagiousness and pathogenicity. These viruses pose a significant threat to fish aquaculture, resulting in substantial annual economic losses for the fish farming industry. Therefore, the development of novel, rapid virus detection technologies is essential for the prevention and control of ISKNV and MRV diseases. In this study, we developed a rapid, sensitive, and visual detection method for MRV and ISKNV using the recombinase polymerase amplification (RPA)-CRISPR/Cas12a system. This method can detect as low as 1 copy/μL of MRV and 0.1 copy/μL of ISKNV, demonstrating excellent specificity and reproducibility. The detection can be performed at a constant temperature of 37-39°C, eliminating the need for complex equipment. A 30-min RPA amplification followed by a 15-min CRISPR/Cas reaction is sufficient for detecting most samples. For low-concentration samples, extending the CRISPR/Cas reaction time to 60 min improves result visibility. The designed RPA reaction system is capable of performing reverse transcription of RNA, allowing for the detection of mRNA transcribed from the MCP gene of MRV and ISKNV in the sample. Furthermore, two probes were identified that can be observed without the need for excitation light. In conclusion, a field-suitable detection method for ISKNV and MRV has been established, providing a powerful tool for the prompt diagnosis of these aquatic pathogens and aiding in the prevention and control of ISKNV and MRV diseases.
Keywords: CRISPR/Cas12a; ISKNV; RPA; iridovirus; ranavirus.
Copyright © 2024 Lu, Liang, Li, Xu, Weng, He and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
The development of RT-RPA and CRISPR-Cas12a based assay for sensitive detection of Hirame novirhabdovirus.Microb Pathog. 2024 Nov;196:106959. doi: 10.1016/j.micpath.2024.106959. Epub 2024 Sep 19. Microb Pathog. 2024. PMID: 39303955
-
Development of cross-priming amplification coupled with vertical flow visualization for rapid detection of infectious spleen and kidney necrosis virus (ISKNV) in mandarin fish, Siniperca chuatsi.J Virol Methods. 2018 Mar;253:38-42. doi: 10.1016/j.jviromet.2017.12.008. Epub 2017 Dec 26. J Virol Methods. 2018. PMID: 29288074
-
Ov-RPA-CRISPR/Cas12a assay for the detection of Opisthorchis viverrini infection in field-collected human feces.Parasit Vectors. 2024 Feb 21;17(1):80. doi: 10.1186/s13071-024-06134-7. Parasit Vectors. 2024. PMID: 38383404 Free PMC article.
-
Rapid and sensitive detection of two fungal pathogens in soybeans using the recombinase polymerase amplification/CRISPR-Cas12a method for potential on-site disease diagnosis.Pest Manag Sci. 2024 Mar;80(3):1168-1181. doi: 10.1002/ps.7847. Epub 2023 Dec 1. Pest Manag Sci. 2024. PMID: 37874890
-
KP177R-based visual assay integrating RPA and CRISPR/Cas12a for the detection of African swine fever virus.Front Immunol. 2024 Apr 9;15:1358960. doi: 10.3389/fimmu.2024.1358960. eCollection 2024. Front Immunol. 2024. PMID: 38655256 Free PMC article.
Cited by
-
Human Riboviruses: A Comprehensive Study.J Mol Evol. 2025 Feb;93(1):11-37. doi: 10.1007/s00239-024-10221-9. Epub 2024 Dec 31. J Mol Evol. 2025. PMID: 39739017 Review.
-
Development of a Real-Time Enzymatic Recombinase Amplification Assay (RT-ERA) and an ERA Combined with a Lateral Flow Dipstick (LFD) Assay (ERA-LFD) for Enteric Microsporidian (Enterospora epinepheli) in Grouper Fishes.Biology (Basel). 2025 Mar 25;14(4):330. doi: 10.3390/biology14040330. Biology (Basel). 2025. PMID: 40282195 Free PMC article.
References
-
- Cao W., Huang B., Xu Q., Xie H., Gao J., Mai X., et al. . (2024). Multiplex qPCR development for the simultaneous and rapid detection of largemouth bass virus and infectious spleen and kidney necrosis virus in aquaculture. J. Virol. Methods 330:115012. doi: 10.1016/j.jviromet.2024.115012 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

