Predicting BRAF Mutations in Cutaneous Melanoma Patients Using Neural Network Analysis
- PMID: 39735251
- PMCID: PMC11671645
- DOI: 10.1155/jskc/3690228
Predicting BRAF Mutations in Cutaneous Melanoma Patients Using Neural Network Analysis
Abstract
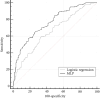
Point mutations at codon 600 of the BRAF oncogene are the most common alterations in cutaneous melanoma (CM). Assessment of BRAF status allows to personalize patient management, though the affordability of molecular testing is limited in some countries. This study aimed to develop a model for predicting alteration in BRAF based on routinely available clinical and histological data. Methods: For identifying the key factors associated with point mutations in BRAF, 2041 patients with CM were recruited in the study. The presence of BRAF mutations was an endpoint. The variables included demographic data (gender and age), anatomic location, stage, histological subtype, number of mitosis, and also such features as ulceration, Clark level, Breslow thickness, infiltration by lymphocytes, invasiveness, regression, microsatellites, and association with nevi. Results: A relatively high rate of BRAF mutation was revealed in the Ukrainian cohort of patients with CM. BRAF-mutant melanoma was associated with younger age and location of nonsun-exposed skin. Besides, sex-specific differences were found between CM of various anatomic distributions and the frequency of distinct BRAF mutation subtypes. A minimal set of variables linked to BRAF mutations, defined by the genetic input selection algorithm, included patient age, primary tumor location, histological type, lymphovascular invasion, ulceration, and association with nevi. To encounter nonlinear links, neural network modeling was applied resulting in a multilayer perceptron (MLP) with one hidden layer. Its architecture included four neurons with a logistic activation function. The AUROCMLP6 of the MLP model comprised 0.79 (95% CІ: 0.74-0.84). Under the optimal threshold, the model demonstrated the following parameters: sensitivity: 89.4% (95% CІ: 84.5%-93.1%), specificity: 50.7% (95% CІ: 42.2%-59.1%), positive predictive value: 73.1% (95% CІ: 69.6%-76.3%), and negative predictive value: 76.0% (95% CІ: 67.6%-82.8%). The developed MLP model enables the prediction of the mutation in BRAF oncogene in CM, alleviating decisions on personalized management of patients with CM. In conclusion, the developed MLP model, which relies on the assessment of 6 variables, can predict the BRAF mutation status in patients with CM, supporting decisions on patient management.
Keywords: BRAF mutation; cutaneous melanoma; multilayered perceptron; predictive model.
Copyright © 2024 Oleksandr Dudin et al.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Incidence of BRAF mutations in cutaneous melanoma: histopathological and molecular analysis of a Ukrainian population.Melanoma Manag. 2023 Dec 21;10(1):MMT64. doi: 10.2217/mmt-2023-0005. eCollection 2023 Mar. Melanoma Manag. 2023. PMID: 38221928 Free PMC article.
-
Defining the high-risk category of patients with cutaneous melanoma: a practical tool based on prognostic modeling.Front Mol Biosci. 2025 Feb 7;12:1543148. doi: 10.3389/fmolb.2025.1543148. eCollection 2025. Front Mol Biosci. 2025. PMID: 39990871 Free PMC article.
-
Russian study of morphological prognostic factors characterization in BRAF-mutant cutaneous melanoma.Pathol Res Pract. 2015 Jul;211(7):521-7. doi: 10.1016/j.prp.2015.03.005. Epub 2015 Apr 2. Pathol Res Pract. 2015. PMID: 25888143
-
Black and Brown Oro-facial Mucocutaneous Neoplasms.Head Neck Pathol. 2019 Mar;13(1):56-70. doi: 10.1007/s12105-019-01008-2. Epub 2019 Jan 29. Head Neck Pathol. 2019. PMID: 30693458 Free PMC article. Review.
-
Defining the Criteria for Reflex Testing for BRAF Mutations in Cutaneous Melanoma Patients.Cancers (Basel). 2021 May 10;13(9):2282. doi: 10.3390/cancers13092282. Cancers (Basel). 2021. PMID: 34068774 Free PMC article. Review.
References
-
- Buckelew H., Crowe C. Social Media and Pathology: A Powerful Intersection for Visibility. American Journal of Clinical Pathology . 2024;162(Supplement_1):S136–S137. doi: 10.1093/AJCP/AQAE129.303. - DOI
LinkOut - more resources
Full Text Sources
Research Materials

