New insights into the putative role of leucine-rich repeat proteins of Leptospira interrogans and their participation in host cell invasion: an in silico analysis
- PMID: 39735260
- PMCID: PMC11674859
- DOI: 10.3389/fcimb.2024.1492352
New insights into the putative role of leucine-rich repeat proteins of Leptospira interrogans and their participation in host cell invasion: an in silico analysis
Abstract
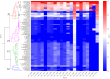
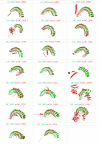
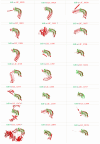
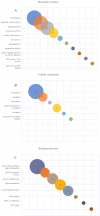
Pathogenic Leptospira are spirochetes that cause leptospirosis, a worldwide zoonotic disease. Leptospirosis affects humans and animals, with approximately 1 million human infections and 60,000 deaths per year. The diversity of leptospiral strains and serovars allied to the fact that pathogenesis is not yet fully understood, make the development of an effective vaccine against leptospirosis a challenge. Outer membrane and secreted proteins are considered potential antigens since they play a vital role in mediating interactions with host molecules. Several domains or motifs have been reported to participate in the leptospiral infection process. Among them, leucine-rich repeat (LRR) proteins have been highlighted as attractive multipurpose proteins, exhibiting a broad spectrum of ligands and having a putative role in bacterial pathogenesis. Indeed, genome annotation of leptospiral species pointed out that LRR proteins are predominant in pathogenic strains, a feature that corroborates this hypothesis. A few LRR proteins of L. santarosai, L. borgpetersenii and L. interrogans have been studied and their possible role in virulence was proposed. Yet, a mechanistic and broad investigation of LRR proteins was not fully performed. In this review, a comprehensive in silico analysis of 21 LRR proteins of L. interrogans was performed in relation to structure, function, dynamics and virulent potential that will contribute to understanding the key role of these domains in the underlying mechanisms of leptospiral infection.
Keywords: LRR proteins; Leptospira; in sílico analysis; leptospirosis; pathogenesis.
Copyright © 2024 Foltran, Gaspar, Silva, Pires, Andrade, Costa, Paixao, Fernandes, Teixeira and Nascimento.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
In silico analysis and functional characterization of a leucine-rich repeat protein of Leptospira interrogans.Int J Med Microbiol. 2024 Sep;316:151633. doi: 10.1016/j.ijmm.2024.151633. Epub 2024 Aug 29. Int J Med Microbiol. 2024. PMID: 39232290
-
Comparative subproteome analysis of three representative Leptospira interrogans vaccine strains reveals cross-reactive antigens and novel virulence determinants.J Proteomics. 2015 Jan 1;112:27-37. doi: 10.1016/j.jprot.2014.08.015. Epub 2014 Sep 6. J Proteomics. 2015. PMID: 25201075
-
Pathogenic Leptospira interrogans exoproteins are primarily involved in heterotrophic processes.Infect Immun. 2015 Aug;83(8):3061-73. doi: 10.1128/IAI.00427-15. Epub 2015 May 18. Infect Immun. 2015. PMID: 25987703 Free PMC article.
-
Pathogenesis of leptospirosis: the influence of genomics.Vet Microbiol. 2011 Nov 21;153(1-2):73-81. doi: 10.1016/j.vetmic.2011.02.055. Epub 2011 Mar 5. Vet Microbiol. 2011. PMID: 21440384 Review.
-
AAA+ Molecular Chaperone ClpB in Leptospira interrogans: Its Role and Significance in Leptospiral Virulence and Pathogenesis of Leptospirosis.Int J Mol Sci. 2020 Sep 11;21(18):6645. doi: 10.3390/ijms21186645. Int J Mol Sci. 2020. PMID: 32932775 Free PMC article. Review.
Cited by
-
Addressing Critical Fungal Pathogens Under a One Health Perspective: Key Insights from the Portuguese Association of Medical Mycology.Mycopathologia. 2025 Aug 16;190(5):73. doi: 10.1007/s11046-025-00981-3. Mycopathologia. 2025. PMID: 40818022 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

