Examining the Relationship between Aptamer Complexity and Molecular Discrimination of a Low-Epitope Target
- PMID: 39735321
- PMCID: PMC11672540
- DOI: 10.1021/acscentsci.4c01377
Examining the Relationship between Aptamer Complexity and Molecular Discrimination of a Low-Epitope Target
Abstract
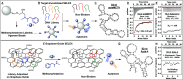
Aptamers are oligonucleotide-based affinity reagents that are increasingly being used in various applications. Systematic evolution of ligands by exponential enrichment (SELEX) has been widely used to isolate aptamers for small-molecule targets, but it remains challenging to generate aptamers with high affinity and specificity for targets with few functional groups. To address this challenge, we have systematically evaluated strategies for optimizing the isolation of aptamers for (+)-methamphetamine, a target for which previously reported aptamers have weak or no binding affinity. We perform four trials of library-immobilized SELEX against (+)-methamphetamine and demonstrate that N30 libraries do not yield high-quality aptamers. However, by using a more complex N40 library design, stringent counter-SELEX, and fine-tuned selection conditions, we identify aptamers with high affinity for (+)-methamphetamine and better selectivity relative to existing antibodies. Bioinformatic analysis from our selections reveals that high-quality aptamers contain long conserved motifs and are more informationally dense. Finally, we demonstrate that our best aptamer can rapidly detect (+)-methamphetamine at toxicologically relevant concentrations in saliva in a colorimetric dye-displacement assay. The insights provided here demonstrate the challenges in generating high-quality aptamers for low complexity small-molecule targets and will help guide the design of more efficient future selection efforts.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures






Similar articles
-
Isolation of Natural DNA Aptamers for Challenging Small-Molecule Targets, Cannabinoids.Anal Chem. 2021 Feb 16;93(6):3172-3180. doi: 10.1021/acs.analchem.0c04592. Epub 2021 Feb 2. Anal Chem. 2021. PMID: 33528997 Free PMC article.
-
Systematic Study of in Vitro Selection Stringency Reveals How To Enrich High-Affinity Aptamers.J Am Chem Soc. 2023 Jan 11;145(1):194-206. doi: 10.1021/jacs.2c09522. Epub 2022 Dec 27. J Am Chem Soc. 2023. PMID: 36574475
-
Rapid Nuclease-Assisted Selection of High-Affinity Small-Molecule Aptamers.J Am Chem Soc. 2024 Aug 7;146(31):21296-21307. doi: 10.1021/jacs.4c00748. Epub 2024 Jul 23. J Am Chem Soc. 2024. PMID: 39042584
-
Selection of aptamers targeting small molecules by capillary electrophoresis: Advances, challenges, and prospects.Biotechnol Adv. 2025 Jan-Feb;78:108491. doi: 10.1016/j.biotechadv.2024.108491. Epub 2024 Nov 26. Biotechnol Adv. 2025. PMID: 39603433 Review.
-
Capture-SELEX: Selection Strategy, Aptamer Identification, and Biosensing Application.Biosensors (Basel). 2022 Dec 7;12(12):1142. doi: 10.3390/bios12121142. Biosensors (Basel). 2022. PMID: 36551109 Free PMC article. Review.
Cited by
-
Highly selective DNA aptamer sensor for intracellular detection of coenzyme A.Chem Sci. 2025 Mar 28;16(18):8023-8029. doi: 10.1039/d5sc00332f. eCollection 2025 May 7. Chem Sci. 2025. PMID: 40206557 Free PMC article.
-
Exploring the relationship between aptamer binding thermodynamics, affinity, and specificity.Nucleic Acids Res. 2025 Mar 20;53(6):gkaf219. doi: 10.1093/nar/gkaf219. Nucleic Acids Res. 2025. PMID: 40156861 Free PMC article.
References
-
- Dunn M. R.; Jimenez R. M.; Chaput J. C. Analysis of Aptamer Discovery and Technology. Nat. Rev. Chem. 2017, 1 (10), 0076.10.1038/s41570-017-0076. - DOI
LinkOut - more resources
Full Text Sources
