Generative dynamical models for classification of rsfMRI data
- PMID: 39735493
- PMCID: PMC11675094
- DOI: 10.1162/netn_a_00412
Generative dynamical models for classification of rsfMRI data
Abstract
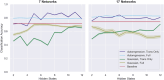
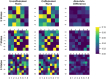
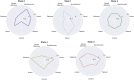
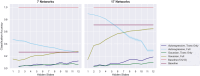
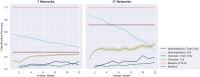
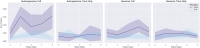
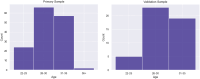
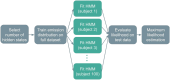
The growing availability of large-scale neuroimaging datasets and user-friendly machine learning tools has led to a recent surge in studies that use fMRI data to predict psychological or behavioral variables. Many such studies classify fMRI data on the basis of static features, but fewer try to leverage brain dynamics for classification. Here, we pilot a generative, dynamical approach for classifying resting-state fMRI (rsfMRI) data. By fitting separate hidden Markov models to the classes in our training data and assigning class labels to test data based on their likelihood under those models, we are able to take advantage of dynamical patterns in the data without confronting the statistical limitations of some other dynamical approaches. Moreover, we demonstrate that hidden Markov models are able to successfully perform within-subject classification on the MyConnectome dataset solely on the basis of transition probabilities among their hidden states. On the other hand, individual Human Connectome Project subjects cannot be identified on the basis of hidden state transition probabilities alone-although a vector autoregressive model does achieve high performance. These results demonstrate a dynamical classification approach for rsfMRI data that shows promising performance, particularly for within-subject classification, and has the potential to afford greater interpretability than other approaches.
Keywords: Classification; Generative models; Hidden Markov models; Network dynamics; Resting-state fMRI.
Plain language summary
Neuroimaging researchers have made substantial progress in using brain data to predict psychological and behavioral variables, like personality, cognitive abilities, and neurological and psychiatric diagnoses. In general, however, these prediction approaches do not take account of how brain activity changes over time. In this study, we use hidden Markov models, a simple and generic model for dynamic processes, to perform brain-based prediction. We show that hidden Markov models can successfully distinguish whether a single individual had eaten and consumed caffeine before his brain scan. These models also show some promise for “fingerprinting,” or identifying individuals solely on the basis of their brain scans. This study demonstrates that hidden Markov models are a promising tool for neuroimaging-based prediction.
© 2024 Massachusetts Institute of Technology.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures






Similar articles
-
Bayesian switching factor analysis for estimating time-varying functional connectivity in fMRI.Neuroimage. 2017 Jul 15;155:271-290. doi: 10.1016/j.neuroimage.2017.02.083. Epub 2017 Mar 4. Neuroimage. 2017. PMID: 28267626 Free PMC article.
-
Predicting individual traits from models of brain dynamics accurately and reliably using the Fisher kernel.Elife. 2025 Jan 31;13:RP95125. doi: 10.7554/eLife.95125. Elife. 2025. PMID: 39887179 Free PMC article.
-
A Novel Hidden Markov Approach to Studying Dynamic Functional Connectivity States in Human Neuroimaging.Brain Connect. 2023 Apr;13(3):154-163. doi: 10.1089/brain.2022.0031. Epub 2023 Feb 16. Brain Connect. 2023. PMID: 36367193 Free PMC article.
-
Interpreting temporal fluctuations in resting-state functional connectivity MRI.Neuroimage. 2017 Dec;163:437-455. doi: 10.1016/j.neuroimage.2017.09.012. Epub 2017 Sep 12. Neuroimage. 2017. PMID: 28916180 Review.
-
Neural and metabolic basis of dynamic resting state fMRI.Neuroimage. 2018 Oct 15;180(Pt B):448-462. doi: 10.1016/j.neuroimage.2017.09.010. Epub 2017 Sep 9. Neuroimage. 2018. PMID: 28899744 Free PMC article. Review.
References
-
- Bondi, E., Maggioni, E., Brambilla, P., & Delvecchio, G. (2023). A systematic review on the potential use of machine learning to classify major depressive disorder from healthy controls using resting state fMRI measures. Neuroscience & Biobehavioral Reviews, 144, 104972. 10.1016/j.neubiorev.2022.104972, - DOI - PubMed
-
- Bustamante, C., Castrillón, G., & Arias-Londoño, J. (2023). Classification of focused perturbations using time-variant functional connectivity with rs-fMRI. In Communications in computer and information science (Vol. 1746, pp. 18–30). Springer. 10.1007/978-3-031-29783-0_2 - DOI
LinkOut - more resources
Full Text Sources
