A generative model of the connectome with dynamic axon growth
- PMID: 39735503
- PMCID: PMC11674315
- DOI: 10.1162/netn_a_00397
A generative model of the connectome with dynamic axon growth
Abstract
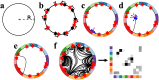
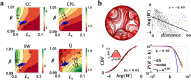
Connectome generative models, otherwise known as generative network models, provide insight into the wiring principles underpinning brain network organization. While these models can approximate numerous statistical properties of empirical networks, they typically fail to explicitly characterize an important contributor to brain organization-axonal growth. Emulating the chemoaffinity-guided axonal growth, we provide a novel generative model in which axons dynamically steer the direction of propagation based on distance-dependent chemoattractive forces acting on their growth cones. This simple dynamic growth mechanism, despite being solely geometry-dependent, is shown to generate axonal fiber bundles with brain-like geometry and features of complex network architecture consistent with the human brain, including lognormally distributed connectivity weights, scale-free nodal degrees, small-worldness, and modularity. We demonstrate that our model parameters can be fitted to individual connectomes, enabling connectome dimensionality reduction and comparison of parameters between groups. Our work offers an opportunity to bridge studies of axon guidance and connectome development, providing new avenues for understanding neural development from a computational perspective.
Keywords: Axon simulation; Connectome; Generative model; Network neuroscience.
Plain language summary
Generative models of the human connectome provide insight into principles driving brain network development. However, current models do not capture axonal outgrowth, which is crucial to the formation of neural circuits. We develop a novel generative connectome model featuring dynamic axonal outgrowth, revealing the contribution of microscopic axonal guidance to the network topology and axonal geometry of macroscopic connectomes. Simple axonal outgrowth rules representing continuous chemoaffinity gradients are shown to generate complex, brain-like topologies and realistic axonal fascicle architectures. Our model is sufficiently sensitive to capture subtle interindividual differences in axonal outgrowth between healthy adults. Our results are significant because they reveal core principles that may give rise to both complex brain networks and brain-like axonal bundles, unifying neurogenesis across scales.
© 2024 Massachusetts Institute of Technology.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures



Update of
-
A generative model of the connectome with dynamic axon growth.bioRxiv [Preprint]. 2024 Feb 28:2024.02.23.581824. doi: 10.1101/2024.02.23.581824. bioRxiv. 2024. Update in: Netw Neurosci. 2024 Dec 10;8(4):1192-1211. doi: 10.1162/netn_a_00397. PMID: 38464116 Free PMC article. Updated. Preprint.
References
LinkOut - more resources
Full Text Sources
