Simulated brain networks reflecting progression of Parkinson's disease
- PMID: 39735513
- PMCID: PMC11675161
- DOI: 10.1162/netn_a_00406
Simulated brain networks reflecting progression of Parkinson's disease
Abstract
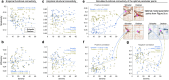
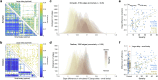
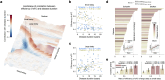
The neurodegenerative progression of Parkinson's disease affects brain structure and function and, concomitantly, alters the topological properties of brain networks. The network alteration accompanied by motor impairment and the duration of the disease has not yet been clearly demonstrated in the disease progression. In this study, we aim to resolve this problem with a modeling approach using the reduced Jansen-Rit model applied to large-scale brain networks derived from cross-sectional MRI data. Optimizing whole-brain simulation models allows us to discover brain networks showing unexplored relationships with clinical variables. We observe that the simulated brain networks exhibit significant differences between healthy controls (n = 51) and patients with Parkinson's disease (n = 60) and strongly correlate with disease severity and disease duration of the patients. Moreover, the modeling results outperform the empirical brain networks in these clinical measures. Consequently, this study demonstrates that utilizing the simulated brain networks provides an enhanced view of network alterations in the progression of motor impairment and identifies potential biomarkers for clinical indices.
Keywords: Brain network; Parkinson’s disease; Progressive disease; Whole-brain model.
Plain language summary
Understanding the progression of neurodegenerative diseases is of extreme importance in medicine. We utilize biophysical whole-brain models to describe how the brain networks change in Parkinson’s disease (PD). We demonstrate clear correlations between the severity of motor impairment and the properties of the simulated brain networks, which are not prominent in empirical brain networks. Furthermore, we show that healthy participants exhibit a pronounced adaptation of network efficiencies in response to varying parameters of the model, while such an adaptation process is suppressed in PD patients with higher disease severity and duration. Our findings suggest a potential model-based biomarker for classification and clinical evaluation of progressive PD using cross-sectional clinical MRI data.
© 2024 Massachusetts Institute of Technology.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures


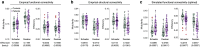
References
-
- Ashburner, J., Friston, K. J., & Penny, W. (2003). Human brain function (2nd ed.). Academic Press. 10.1016/B978-0-12-264841-0.X5000-8 - DOI
-
- Benjamini, Y., & Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological), 57(1), 289–300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
LinkOut - more resources
Full Text Sources
