Development of a urine-based metabolomics approach for multi-cancer screening and tumor origin prediction
- PMID: 39735533
- PMCID: PMC11671364
- DOI: 10.3389/fimmu.2024.1449103
Development of a urine-based metabolomics approach for multi-cancer screening and tumor origin prediction
Abstract
Background: Cancer remains a leading cause of mortality worldwide. A non-invasive screening solution was required for early diagnosis of cancer. Multi-cancer early detection (MCED) tests have been considered to address the challenge by simultaneously identifying multiple types of cancer within a single test using minimally invasive blood samples. However, a multi-cancer screening strategy utilizing urine-based metabolomics has not yet been developed.
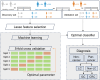
Methods: We enrolled 911 cancer patients with 548 lung cancer (LC), 177 with gastric cancer (GC), and 186 with colorectal cancer (CRC), alongside 563 individuals with non-cancerous benign diseases and 229 healthy controls (HC) and investigated the metabolic profiles of urine samples. Participants were randomly allocated to discovery and validation cohorts. The discovery cohort was used for identifying multi-cancer and tissue-specific signatures to build the cancer screening and tumor origin prediction models, while the validation cohort was employed for assessing the performance of these models.
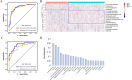
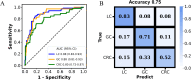
Results: We identified and annotated a total of 360 metabolites from the urine samples. Using the LASSO regression algorithm, 18 metabolites were characterized as urinary metabolic biomarkers and exhibited excellent discriminative performance between cancer patients and HC with AUC of 0.96 in the validation cohort. In comparison with the performance of traditional tumor markers CEA, the screening model performed higher sensitivity across the cancer stages, with a particularly increase in sensitivity among early-stage cancer patients. Moreover, the screening model also exhibited in high classification of cancers from non-cancerous group, comprising with HC and benign disease participants. Furthermore, two non-overlapping metabolic panels were selected to differentiate LC from Non-LC and GC from CRC with the AUC values of 0.87 and 0.83 in validation cohorts, respectively. Additionally, the model accurately predicted the origin of three lethal cancers: lung, gastric, and colorectal, with an overall accuracy of 0.75. The AUC values for LC, GC, and CRC were 0.88, 0.88, and 0.80, respectively.
Discussion: Our study demonstrates the potential of urine-based metabolomics for multi-cancer early detection. The approach offers non-invasive cancer screening, promising widespread implementation in population-based programs for early detection and improved outcomes. Further validation and expansion are needed for broader clinical applicability.
Keywords: machine learning; multi-cancer screening; pathway; tumor origin prediction; urinary metabolomics.
Copyright © 2024 Xu, Zeng, Qing, He, Song, Wang, Yu, Zhang, Wei, Liu, Wen, Hu, Zhang, Li, Chen and Xia.
Conflict of interest statement
YH, JW, SY, TZ, QW, LL, HW, JH, and YL are employees of Metanotitia Inc., Harbin. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Urine-NMR metabolomics for screening of advanced colorectal adenoma and early stage colorectal cancer.Sci Rep. 2019 Mar 18;9(1):4786. doi: 10.1038/s41598-019-41216-y. Sci Rep. 2019. PMID: 30886205 Free PMC article.
-
Identification of Diagnostic Biomarkers for Colorectal Polyps Based on Noninvasive Urinary Metabolite Screening and Construction of a Nomogram.Cancer Med. 2025 Apr;14(7):e70762. doi: 10.1002/cam4.70762. Cancer Med. 2025. PMID: 40200572 Free PMC article.
-
Urinary Metabolomics to Identify a Unique Biomarker Panel for Detecting Colorectal Cancer: A Multicenter Study.Cancer Epidemiol Biomarkers Prev. 2019 Aug;28(8):1283-1291. doi: 10.1158/1055-9965.EPI-18-1291. Epub 2019 May 31. Cancer Epidemiol Biomarkers Prev. 2019. PMID: 31151939 Free PMC article.
-
Multi-cancer early detection tests for general population screening: a systematic literature review.Health Technol Assess. 2025 Jan;29(2):1-105. doi: 10.3310/DLMT1294. Health Technol Assess. 2025. PMID: 39898371 Free PMC article.
-
Cancer metabolomic markers in urine: evidence, techniques and recommendations.Nat Rev Urol. 2019 Jun;16(6):339-362. doi: 10.1038/s41585-019-0185-3. Nat Rev Urol. 2019. PMID: 31092915 Review.
Cited by
-
Metabolomic Profiling of Disease Progression Following Radiotherapy for Breast Cancer.Cancers (Basel). 2025 Mar 5;17(5):891. doi: 10.3390/cancers17050891. Cancers (Basel). 2025. PMID: 40075737 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

