Identification and verification of the optimal feature genes of ferroptosis in thyroid-associated orbitopathy
- PMID: 39735537
- PMCID: PMC11671519
- DOI: 10.3389/fimmu.2024.1422497
Identification and verification of the optimal feature genes of ferroptosis in thyroid-associated orbitopathy
Abstract
Background: Thyroid-associated orbitopathy (TAO) is an autoimmune inflammatory disorder of the orbital adipose tissue, primarily causing oxidative stress injury and tissue remodeling in the orbital connective tissue. Ferroptosis is a form of programmed cell death driven by the accumulation of reactive oxygen species (ROS), iron metabolism disorder, and lipid peroxidation. This study aims to identify and validate the optimal feature genes (OFGs) of ferroptosis with diagnostic and therapeutic potential in TAO orbital adipose tissue through bioinformatics analysis and to assess their correlation with disease-related immune cell infiltration.
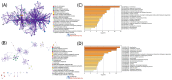
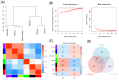
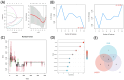
Methods: Search of the Gene Expression Omnibus database for TAO-related gene datasets led to the selection of GSE58331 for differential gene expression analysis. WGCNA was employed to identify key disease modules and hub genes. The intersection of DEGs, hub genes and ferroptosis-related gene yielded key genes of ferroptosis. Machine learning algorithms identified OFGs of ferroptosis. Meanwhile, by comparing the expression of FRGs in the orbital adipose tissue and the orbital fibroblasts (OFs) of healthy controls and TAO patients, as well as co-culturing macrophages and OFs in vitro, the influence of macrophages on FRGs in OFs was explored. CIBERSORT analyzed immune cell infiltration to determine proportions of immune cell types in each sample, and Spearman correlation analysis explored relationships between OFGs and infiltrating immune cells. Finally, GSEA determined the function of each key biomarker based on the median expression of OFGs.
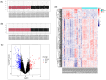
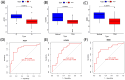
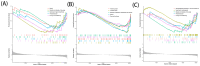
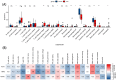
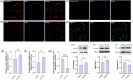
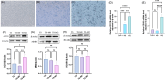
Results: Three TAO FRGs (ACO1, MMD, and HCAR1) were screened in the dataset. The ROC results of ACO1 showed that the AUC value was greater than 0.8 in all the datasets, which was the strongest for disease specificity and diagnostic ability. Validation results showed that, in addition to MMD, the expression of ACO1 and HCAR1 in orbital adipose tissue of TAO patients was significantly down-regulated, while M2-type macrophages might be involved in regulating the expression of ACO1 in orbital adipose-derived OFs. CIBERSORT immune cell infiltration analysis showed that in orbital adipose tissue of TAO patients, memory B-lymphocytes, T regulatory cells, NK-cells, M0-type macrophages, M1-type macrophages, resting dendritic cells, activated mast cells, and neutrophils infiltration levels were significantly elevated.
Conclusion: Through bioinformatics analysis, this study identified and validated two OFGs of ferroptosis with diagnostic and therapeutic potential in TAO orbital adipose tissue, suggesting that the downregulation of ACO1 and HCAR1 may be potential molecular targets in the pathogenesis of TAO.
Keywords: GEO; WGCNA; ferroptosis-related gene; immune cell infiltration; orbitopathy.
Copyright © 2024 Li, Xiong, Wang, Ren, Jin, Yu, Chen, Gan, Xu, Wang and Liao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

