Long noncoding RNA LINC01106 promotes lung adenocarcinoma progression via upregulation of autophagy
- PMID: 39735666
- PMCID: PMC11671404
- DOI: 10.32604/or.2024.047626
Long noncoding RNA LINC01106 promotes lung adenocarcinoma progression via upregulation of autophagy
Abstract
Background: Long noncoding RNA, LINC01106 exhibits high expression in lung adenocarcinoma (LUAD) tumor tissues, but its functional role and regulatory mechanism in LUAD cells remain unclear.
Methods: LINC01106 expression was analyzed in LUAD tissues and its functional impact on LUAD cells was assessed. LUAD cells were silenced with sh-LINC01106 and injected into nude mice to investigate tumor growth. The downstream transcription factors and molecular mechanism were determined using the Human transcription factor database (TFDB) database and Gene Expression Profiling Interactive Analysis (GEPIA) database. Additionally, the impact of linc01106 on autophagy was analyzed by determining the expression of autophagy-related genes (ATGs) in LUAD cells.
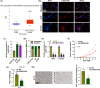
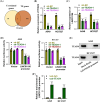
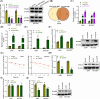
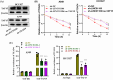
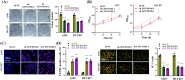
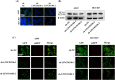
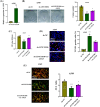
Results: Our results showed that LINC01106 exhibited upregulation in both LUAD tissues and cell lines. The silencing of LINC01106 demonstrated a suppressive effect on tumorigenesis in a xenograft mouse model of LUAD. Additionally, LINC01106 was found to recruit TATA-binding protein-associated factor 15 (TAF15), an RNA-binding protein, thereby enhancing the mRNA stability of TEA domain transcription factor 4 (TEAD4). In turn, TEAD4 served as a transcription factor that bound to the LINC01106 promoter and regulated its expression. Further assays indicated that LINC01106 promoted autophagy in LUAD cells by upregulating the expression of autophagy-related genes (ATGs). The silencing of LINC01106 in LUAD cells inhibited autophagy, and cell proliferation, and promoted apoptosis, which all were effectively reversed by ATG5 overexpression.
Conclusions: Overall, LINC01106, transcriptionally activated by TEAD4, interacts with TAF15 to promote the stability of TEAD4 and upregulates the expression of ATGs, promoting malignancy of LUAD cells.
Keywords: ATG5; LINC01106; Lung adenocarcinoma (LUAD); Non-small cell lung cancer (NSCLC); TAF15; TEAD4.
© 2025 The Authors.
Conflict of interest statement
The authors declare no conflicts of interest to report regarding the present study.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
