Identification and analysis of major latex protein (MLP) family genes in Rosa chinensis responsive to Botrytis cinerea infection by RNA-seq approaches
- PMID: 39735770
- PMCID: PMC11671256
- DOI: 10.3389/fpls.2024.1511597
Identification and analysis of major latex protein (MLP) family genes in Rosa chinensis responsive to Botrytis cinerea infection by RNA-seq approaches
Abstract
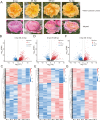
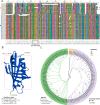
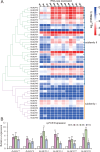
Roses (Rosa chinensis) are among the most cherished ornamental plants globally, yet they are highly susceptible to infections by Botrytis cinerea, the causative agent of gray mold disease. Here we inoculated the resistant rose variety 'Yellow Leisure Liness' with B. cinerea to investigate its resistance mechanisms against gray mold disease. Through transcriptome sequencing, we identified 578 differentially expressed genes (DEGs) that were significantly upregulated at 24, 48, and 72 hours post-inoculation, with these genes significantly enriched for three defense response-related GO terms. Further domain analysis of the genes in these GO terms reveal that 21 DEGs contain the Bet v 1 family domain, belonging to the major latex protein (MLP) gene family, suggesting their potential key role in rose disease resistance. Furthermore, we systematically identified 46 RcMLP genes in roses and phylogenetically categorized them into two distinct subfamilies: group I and II. Genomic duplication analysis indicates that tandem duplication is the main driver for the expansion of the RcMLP family, and these genes have undergone by purifying selection. Additionally, detailed analyses of gene structure, motif composition, and promoter regions reveal that RcMLP genes contain numerous stress-responsive elements, with 32 RcMLP genes harboring fungal elicitor/wound-responsive elements. The constructed potential transcription factor regulatory network showed significant enrichment of the ERF transcription factor family in the regulation of RcMLP genes. Gene expression analysis reveal that DEGs are mainly distributed in subfamily II, where four highly expressed genes (RcMLP13, RcMLP28, RcMLP14, and RcMLP27) are identified in a small branch, with their fold change exceeding ten folds and verified by qRT-PCR. In summary, our research results underscore the potential importance of the RcMLP gene family in response to B. cinerea infection and provide comprehensive basis for further function exploration of the MLP gene family in rose resistance to fungal infections.
Keywords: Botrytis cinerea; RNA-Seq; major latex protein; rose; tandem duplication.
Copyright © 2024 Chen, Li, Cheng, Yan, Dong, Hou, Zhu and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Comparative RNA-Seq analysis reveals a critical role for brassinosteroids in rose (Rosa hybrida) petal defense against Botrytis cinerea infection.BMC Genet. 2018 Aug 20;19(1):62. doi: 10.1186/s12863-018-0668-x. BMC Genet. 2018. PMID: 30126371 Free PMC article.
-
Global analysis of the AP2/ERF gene family in rose (Rosa chinensis) genome unveils the role of RcERF099 in Botrytis resistance.BMC Plant Biol. 2020 Nov 23;20(1):533. doi: 10.1186/s12870-020-02740-6. BMC Plant Biol. 2020. PMID: 33228522 Free PMC article.
-
Genome-wide characterization of the rose (Rosa chinensis) WRKY family and role of RcWRKY41 in gray mold resistance.BMC Plant Biol. 2019 Nov 27;19(1):522. doi: 10.1186/s12870-019-2139-6. BMC Plant Biol. 2019. PMID: 31775626 Free PMC article.
-
Comprehensive analysis of bZIP gene family and function of RcbZIP17 on Botrytis resistance in rose (Rosa chinensis).Gene. 2023 Jan 15;849:146867. doi: 10.1016/j.gene.2022.146867. Epub 2022 Sep 14. Gene. 2023. PMID: 36115481
-
Disease resistance breeding in rose: current status and potential of biotechnological tools.Plant Sci. 2014 Nov;228:107-17. doi: 10.1016/j.plantsci.2014.04.005. Epub 2014 Apr 13. Plant Sci. 2014. PMID: 25438791 Review.
References
LinkOut - more resources
Full Text Sources

