Sequencing and Optical Genome Mapping for the Adventurous Chemist
- PMID: 39735829
- PMCID: PMC11673194
- DOI: 10.1021/cbmi.4c00060
Sequencing and Optical Genome Mapping for the Adventurous Chemist
Abstract
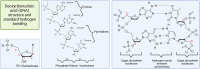
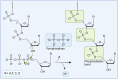
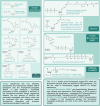
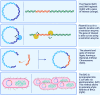
This review provides a comprehensive overview of the chemistries and workflows of the sequencing methods that have been or are currently commercially available, providing a very brief historical introduction to each method. The main optical genome mapping approaches are introduced in the same manner, although only a subset of these are or have ever been commercially available. The review comes with a deck of slides containing all of the figures for ease of access and consultation.
© 2024 The Authors. Co-published by Nanjing University and American Chemical Society.
Conflict of interest statement
The authors declare the following competing financial interest(s): Johan Hofkens and Volker Leen are co-founders of Perseus Biomics.
Figures

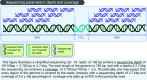



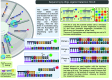










References
-
- Alberts B.; et al. DNA Replication Mechanisms. Molecular Biology of the Cell, 4th ed.; Garland Science, 2002.
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
