Single-Cell Transcriptome Reveals the Heterogeneity of T Cells in Mice with Systemic Lupus Erythematosus and Neuronal Inflammation
- PMID: 39735894
- PMCID: PMC11675326
- DOI: 10.2147/JIR.S474211
Single-Cell Transcriptome Reveals the Heterogeneity of T Cells in Mice with Systemic Lupus Erythematosus and Neuronal Inflammation
Abstract
Introduction: Systemic lupus erythematosus is a heterogeneous autoimmune disease. A burst of autoimmune reactions in various systems can lead to severe clinical conditions closely associated with mortality. T cells serve as mediators that drive the occurrence and maintenance of inflammatory processes.
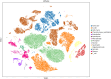
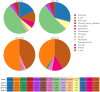
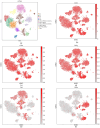
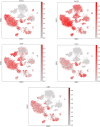
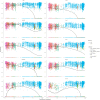
Methods: In this work, we employed single-cell transcriptome sequencing (scRNA-seq) involving 27704 cells from the brain and spleen tissues of MRL/lpr mice and 25355 healthy controls from BALB/c mice to explore the heterogeneity of T cells and their migration from the spleen to the brain.
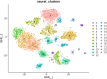
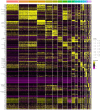
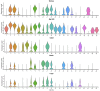
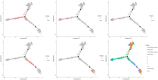
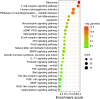
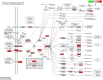
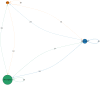
Results: We identified a distinct group of double-negative T cells in systemic lupus erythematosus (SLE) mice that significantly expressed Eomes and other specific markers. We used the analysis of pseudotime trajectory and enrichment to show that double-negative T cells in SLE mice are strongly associated with cellular senescence and exhaustion. Additionally, we focused on the interactions among DNT, astrocytes, and microglia in the mice brain. We observed greater expression of MDK-related ligand‒receptor pairs between astrocytes and double-negative T cells, indicating that MDK may be a therapeutic target for treating neuroinflammation in SLE.
Discussion: This research sheds light on the intricate dynamics of immune responses in mice with SLE, specifically highlighting the role of double-negative T cells and their connection to cellular senescence. The exploration of interactions between T cells, astrocytes, and microglia in the mice brain unveils potential avenues for therapeutic intervention, particularly in addressing neuronal inflammation in SLE.
Keywords: SLE; T cell; neuroinflammation; single-cell transcriptome.
© 2024 Shi et al.
Conflict of interest statement
The authors declare no conflict of interest.
Figures











References
LinkOut - more resources
Full Text Sources

