Combined Blockade of Lipid Uptake and Synthesis by CD36 Inhibitor and SCD1 siRNA Is Beneficial for the Treatment of Refractory Prostate Cancer
- PMID: 39736148
- PMCID: PMC11848597
- DOI: 10.1002/advs.202412244
Combined Blockade of Lipid Uptake and Synthesis by CD36 Inhibitor and SCD1 siRNA Is Beneficial for the Treatment of Refractory Prostate Cancer
Abstract
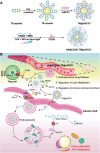
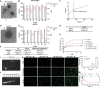
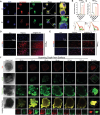
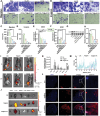
Drug resistance is an important factor for prostate cancer (PCa) to progress into refractory PCa, and abnormal lipid metabolism usually occurs in refractory PCa, which presents great challenges for PCa therapy. Here, a cluster of differentiation 36 (CD36) inhibitor sulfosuccinimidyl oleate sodium (CD36i) and stearoyl-CoA desaturase 1 (SCD1) siRNA (siSCD1) are selected to inhibit lipid uptake and synthesis in PCa, respectively. To this end, a multiresponsive drug delivery nanosystem, HA@CD36i-TR@siSCD1 is designed. The hyaluronic acid (HA) gel "shell" of HA-TR nanosystem can release drugs in response to the acidic tumor microenvironment and hyaluronidase, and the tumor targeting (TR) cationic micellar "core" can release drugs in response to glutathione. This multiresponsive drug release is beneficial for the exogenous inhibition of lipid uptake by CD36i and the endogenous inhibition of lipid synthesis by siSCD1. The established HA-TR nanosystem has good tumor targeting ability and tumor penetration ability, and that HA@CD36i-TR@siSCD1 has good synergistic effects, which can significantly restrain the growth, invasion, and metastasis of PCa. Moreover, under high-fat conditions, the tumors are more sensitive to HA@CD36i-TR@siSCD1 treatment, almost no accumulation of lipid droplets is observed in HA@CD36i-TR@siSCD1-treated tumors, with enhanced antitumor immunity. Hence, this study provides a new treatment option for refractory PCa patients, especially those with a high-fat diet.
Keywords: CD36; SCD1; drug resistance; lipid metabolism; refractory prostate cancer.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

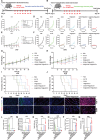
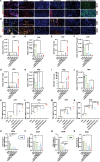

References
-
- Bray F., Laversanne M., Sung H., Ferlay J., Siegel R. L., Soerjomataram I., Jemal A., CA Cancer J. Clin. 2024, 74, 229. - PubMed
-
- Siegel R. L., Giaquinto A. N., Jemal A., CA Cancer J. Clin. 2024, 74, 12. - PubMed
-
- Maluf F. C., Pereira F. M. T., Silva A. G., Dettino A. L. A., Cardoso A. P. G., Sasse A. S., Soares A., Kann A. G., Herchenhorn D., Jardim D. L. F., Cortés D. E. L., Kater F. R., Morbeck I. A. P., Reolon J. F. N., Rinck J. A. J., Zarbá J. J., Sade J. P., da Trindade K. M., Costa L. A. G. A., Dos Santos L. V., Maia M. C., Siqueira M. B., Gillessen S., JCO Glob. Oncol. 2021, 7, 559. - PMC - PubMed
-
- Schaeffer E. M., Srinivas S., Adra N., An Y., Barocas D., Bitting R., Bryce A., Chapin B., Cheng H. H., D'Amico A. V., Desai N., Dorff T., Eastham J. A., Farrington T. A., Gao X., Gupta S., Guzzo T., Ippolito J. E., Kuettel M. R., Lang J. M., Lotan T., McKay R. R., Morgan T., Netto G., Pow‐Sang J. M., Reiter R., Roach M., Robin T., Rosenfeld S., Shabsigh A., et al., J. Natl. Compr. Canc. Netw. 2023, 21, 1067. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
