The therapeutic potential of RNA m(6)A in lung cancer
- PMID: 39736743
- PMCID: PMC11687105
- DOI: 10.1186/s12964-024-01980-5
The therapeutic potential of RNA m(6)A in lung cancer
Abstract
Lung cancer (LC) is a highly malignant and metastatic form of cancer. The global incidence of and mortality from LC is steadily increasing; the mean 5-year overall survival (OS) rate for LC is less than 20%. This frustrating situation may be attributed to the fact that the pathogenesis of LC remains poorly understood and there is still no cure for mid to advanced LC. Methylation at the N6-position of adenosine (N6mA) of RNA (m(6)A) is widely present in human tissues and organs, and has been found to be necessary for cell development and maintenance of homeostasis. However, numerous basic and clinical studies have demonstrated that RNA m(6)A is deregulated in many human malignancies including LC. This can drive LC malignant characteristics such as proliferation, stemness, invasion, epithelial-mesenchymal transition (EMT), metastasis, and therapeutic resistance. Intriguingly, an increasing number of studies have also shown that eliminating RNA m(6)A dysfunction can exert significant anti-cancer effects on LC such as suppression of cell proliferation and viability, induction of cell death, and reversal of treatment insensitivity. The current review comprehensively discusses the therapeutic potential of RNA m(6)A and its underlying molecular mechanisms in LC, providing useful information for the development of novel LC treatment strategies.
Keywords: Epigenetics; Lung cancer; M(6)A; Target therapy; Tumorigenesis.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethical approval and consent to participate: Not applicable. Consent for publication: All authors consent to publication. Competing interests: The authors declare no competing interests.
Figures
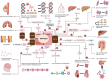
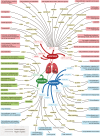
References
-
- Bailey-Wilson JE, Amos CI, Pinney SM, Petersen GM, de Andrade M, Wiest JS, Fain P, Schwartz AG, You M, Franklin W, Klein C, Gazdar A, Rothschild H, Mandal D, Coons T, Slusser J, Lee J, Gaba C, Kupert E, Perez A, Zhou X, Zeng D, Liu Q, Zhang Q, Seminara D, Minna J, Anderson MW. A major lung cancer susceptibility locus maps to chromosome 6q23-25. Am J Hum Genet. 2004;75(3):460–74. - DOI - PMC - PubMed
-
- Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai YY, Chen WV, Shete S, Spitz MR, Houlston RS. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–22. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

