Identification of serum biomarkers for cystic echinococcosis in sheep through untargeted metabolomic analysis using LC-MS/MS technology
- PMID: 39736799
- PMCID: PMC11686990
- DOI: 10.1186/s13071-024-06599-6
Identification of serum biomarkers for cystic echinococcosis in sheep through untargeted metabolomic analysis using LC-MS/MS technology
Abstract
Background: Echinococcosis is a zoonotic disease caused by an Echinococcus tapeworm infection. While diagnostic methods for humans often rely on ultrasound imaging and immunodiagnostic techniques, diagnosis in intermediate hosts typically has no widely used diagnostic markers, hampering disease control efforts.
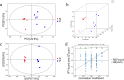
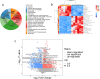
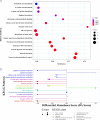
Methods: The differences in serum metabolites of sheep infected with Echinococcus granulosus and a control group were analyzed using ultrahigh-performance liquid chromatography (UHPLC) separation with tandem mass spectrometry (MS/MS) detection. This provided a basis for the early diagnosis and pathogenetic study of cystic echinococcosis (CE) in intermediate hosts at the metabolomics level. Orthogonal projections to latent structures-discriminant analysis (OPLS-DA) were used to analyze different metabolites in the serum of the two groups. The differentially abundant metabolites were entered into the MetaboAnalyst 5.0 online analysis website for processing, and the top-15-ranked metabolic pathways were set to produce bubble plots and differential abundance score plots, with a significant difference of P < 0.05 and a false discovery rate (FDR) < 0.1 as the screening conditions.
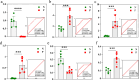
Results: Data analyses of serum samples from both groups identified a total of 1905 significantly different metabolites, where 841 metabolites were upregulated and 1064 metabolites were downregulated. Twelve metabolites were significantly upregulated and 21 metabolites were significantly downregulated in the experimental group. Then, the 1,7-dihydroxyxanthone, 2-methylbutyrylglycine, 3,3-dimethylglutaric acid, 5,12-dihydroxy-6,8,10,14,17-eicosapentaenoic acid, 9-hydroperoxy-10E,12Z,15Z-octadecatrienoic acid, and trimethylamine N-oxide 6 metabolites were selected as diagnostically valuable candidate biomarkers (area under the curve [AUC] > 0.7). These differential metabolites are involved in various metabolic pathways, including amino acid metabolites (arginine, L-isoleucine, L-valine) and fatty acid metabolism (fenugreek, arachidonic acid, linolenic acid). Compared with the control group, sheep in the CE group had increased serum levels of fenugreek acid, while all other metabolites such as glycine showed significantly reduced serum levels (P < 0.01).
Conclusions: Through non-targeted metabolomic analysis of the serum of CE-infected sheep, differential metabolites closely related to amino acid metabolism and the fatty acid metabolism pathway were identified. These differentially abundant metabolites can serve as biomarkers for diagnosing CE infection in intermediate sheep hosts.
Keywords: Echinococcus granulosus; Biomarkers; LC–MS/MS; Non-targeted metabolomics; Serum.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study has been approved by the Institutional Animal Care and Use Committee of Jilin University, China. The approval was granted on September 20, 2022, and this study was registered under the number KT202103004. The current study complies with all relevant laws and international ethics guidelines. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
-
- Casulli A, Abela-Ridder B, Petrone D, Fabiani M, Bobić B, Carmena D, et al. Unveiling the incidences and trends of the neglected zoonosis cystic echinococcosis in Europe: a systematic review from the MEmE project. Lancet Infect Dis. 2023. 10.1016/s1473-3099(22)00638-7. - PubMed
-
- Thompson RC. Biology and systematics of echinococcus. Adv Parasitol. 2017. 10.1016/bs.apar.2016.07.001. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

