Response Surface Methodology for Optimization of Media Components for Production of Lipase from Bacillus subtilis KUBT4
- PMID: 39736954
- PMCID: PMC11682502
- DOI: 10.32592/ARI.2024.79.3.659
Response Surface Methodology for Optimization of Media Components for Production of Lipase from Bacillus subtilis KUBT4
Abstract
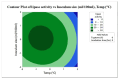
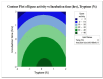
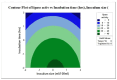
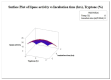
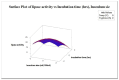
Lipases are triacylglycerol hydrolases with various potential applications because of their different physical properties. Most lipase producers are extracellular in nature and are created using solid-state fermentation and submerged fermentation methods. The fungal, mycelial, and yeast lipases are produced using various solid substrates through the solid-state fermentation method. This method is cost-effective and widely used in industries to produce lipase using fungi. However, lipases from bacteria are produced using submerged fermentation. The optimization of media is a main requirement for increasing the quantitative yield by the overproduction of enzymes. The optimization of media is a main requirement for increasing the quantitative yield by overproduction of enzymes. Different parameters, such as pH, temperature, agitation speed, inoculum size, incubation time, and carbon and nitrogen sources, have been of great importance for researchers in designing economical media. The optimization by one factor at a time (OFAT) is a one-dimensional approach that is laborious and time-consuming and does not consider interactions between the factors. The limitations of OFAT method can be alleviated by employing some techniques, such as Plackett-Burman design (PBD) and response surface methodology (RSM). The PBD is a method to screen the variables that influence production and remove the non-significant factors to attain a smaller and manageable set of factors. Subsequently, the chosen significant factors are optimized by RSM that assists to study the interactions of different factors. The RSM comprises of central composite design (CCD) to fit a second-order polynomial equation. In this study, the effect of temperature, tryptone, inoculum size, and incubation time on the lipase production were analysed by PBD screening experiments. The experiments were designed using a CCD with four variables as part of RSM, utilizing the Design Expert software. This model predicted optimal activity of lipase at 58.53 U/mL when using 1.5% tryptone, a 10 mL inoculum size, and an incubation period of 48 h at 34°C. This experiment was further validated and optimal activity of lipase of 57.85 U/mL was observed. Thus, RSM model enhanced the production of lipase and can be applied for the maximum yield of lipase.
Keywords: Lipase; Plackett-Burman design; Response Surface Methodology; Bacillus subtilis KUBT4.
Conflict of interest statement
Rubeen D. N., Parveen D. N., Makhadumsab M. T., and Shivasharana C. T. declare that they have no conflicts of interest.
Figures








References
-
- Treichel H, De Oliveira D, Mazutti MA, Di Luccio M, Oliveira JV. A review on microbial lipases production. Food Bioproc Tech. 2010;3:182–96.
-
- Nadaf RD, Thimmappa DS. Optimization of the media components by one factor at a time methodology to enhance lipase production by Bacillus Substilis KUBT4. Int J Sci Technol Res. 2020;9(01)
-
- Palaniyappan M, Vijayagopal V, Viswanathan R, Viruthagiri T. Screening of natural substrates and optimization of operating variables on the production of pectinase by submerged fermentation using Aspergillus niger MTCC 281. Afr J Biotechnol. 2009;8(4)
-
- Montgomery DC, Keats JB, Perry LA, Thompson JR, Messina WS. Using statistically designed experiments for process development and improvement: an application in electronics manufacturing. Robot Comput Integr Manuf. 2000;16(1):55–63.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
