Whole-genome sequencing and genomic analysis of four Akkermansia strains newly isolated from human feces
- PMID: 39736996
- PMCID: PMC11683593
- DOI: 10.3389/fmicb.2024.1500886
Whole-genome sequencing and genomic analysis of four Akkermansia strains newly isolated from human feces
Abstract
Background: Numerous studies have demonstrated that Akkermansia is closely associated with human health. These bacteria colonize the mucus layer of the gastrointestinal tract and utilize mucin as their sole source of carbon and nitrogen. Akkermansia spp. exhibit potential as probiotics under specific conditions. However, the gene accumulation curve derived from pan-genome analysis suggests that the genome of Akkermansia strains remains open. Consequently, current genome mining efforts are insufficient to fully capture the intraspecific and interspecific characteristics of Akkermansia, necessitating continuous exploration of the genomic and phenotypic diversity of new isolates.
Methods: Based on this finding, we sequenced, assembled, and functionally annotated the whole genomes of four new human isolates from our laboratory: AKK-HX001, AKK-HX002, AKK-HX003, and AKK-HX004.
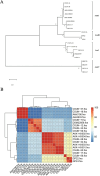
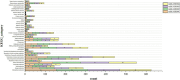
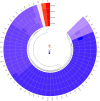
Results: Phylogenetic analysis revealed that all four isolates belonged to the AmII phylogroup, whereas the type strain DSM 22959 is classified within the AmI phylogroup. Moreover, 2,184 shared homologous genes were identified among the four isolates. Functional annotation using the COG, KEGG, and CAZy databases indicated that the functional genes of the four isolates were primarily associated with metabolism. Two antibiotic resistance genes were identified in AKK-HX001 and AKK-HX002, while three resistance genes were detected in AKK-HX003 and AKK-HX004. Additionally, each of the four isolates possessed two virulence genes and three pathogenicity genes, none of which were associated with pathogenicity. The prediction of mobile genetic elements indicated unequal distributions of GIs among the isolates, and a complete CRISPR system was identified in all isolates except AKK-HX003. Two annotated regions of secondary metabolite biosynthesis genes, both belonging to Terpene, were detected using the antiSMASH online tool.
Conclusion: These findings indicate that the four Akkermansia isolates, which belong to a phylogroup distinct from the model strain DSM 22959, exhibit lower genetic risk and may serve as potential probiotic resources for future research.
Keywords: Akkermansia; genomic analysis; genomic diversity; probiotics; whole-genome sequencing.
Copyright © 2024 Lu, Zha, Lyu, LingHu, Chen, Deng, Zhang, Li and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Genome- and Toxicology-Based Safety Assessment of Probiotic Akkermansia muciniphila ONE Isolated from Humans.Foods. 2024 Jun 24;13(13):1979. doi: 10.3390/foods13131979. Foods. 2024. PMID: 38998485 Free PMC article.
-
Akkermansia muciniphila isolated from forest musk deer ameliorates diarrhea in mice via modification of gut microbiota.Animal Model Exp Med. 2025 Feb;8(2):295-306. doi: 10.1002/ame2.12441. Epub 2024 Jun 3. Animal Model Exp Med. 2025. PMID: 38828754 Free PMC article.
-
Comparative Genomics Guides Elucidation of Vitamin B12 Biosynthesis in Novel Human-Associated Akkermansia Strains.Appl Environ Microbiol. 2020 Jan 21;86(3):e02117-19. doi: 10.1128/AEM.02117-19. Print 2020 Jan 21. Appl Environ Microbiol. 2020. PMID: 31757822 Free PMC article.
-
The combination of Clostridium butyricum and Akkermansia muciniphila mitigates DSS-induced colitis and attenuates colitis-associated tumorigenesis by modulating gut microbiota and reducing CD8+ T cells in mice.mSystems. 2025 Feb 18;10(2):e0156724. doi: 10.1128/msystems.01567-24. Epub 2025 Jan 22. mSystems. 2025. PMID: 39840995 Free PMC article.
-
Comparative genomic and functional analysis of Akkermansia muciniphila and closely related species.Genes Genomics. 2019 Nov;41(11):1253-1264. doi: 10.1007/s13258-019-00855-1. Epub 2019 Aug 9. Genes Genomics. 2019. PMID: 31399846 Free PMC article.
References
-
- Alcock B. P., Huynh W., Chalil R., Smith K. W., Raphenya A. R., Wlodarski M. A., et al. . (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 51, D690–D699. doi: 10.1093/nar/gkac920, PMID: - DOI - PMC - PubMed
-
- Al-Emran H. M., Moon J. F., Miah M. L., Meghla N. S., Reuben R. C., Uddin M. J., et al. . (2022). Genomic analysis and in vivo efficacy of Pediococcus acidilactici as a potential probiotic to prevent hyperglycemia, hypercholesterolemia and gastrointestinal infections. Sci. Rep. 12:20429. doi: 10.1038/s41598-022-24791-5, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

