Kv3-Expressing Cells Present More Elaborate N-Glycans with Changes in Cytoskeletal Proteins, Neurite Structure and Cell Migration
- PMID: 39736999
- PMCID: PMC11684427
Kv3-Expressing Cells Present More Elaborate N-Glycans with Changes in Cytoskeletal Proteins, Neurite Structure and Cell Migration
Abstract
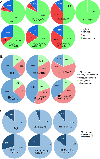
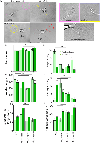
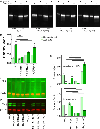
The cues contained by N-glycans relay the quality, cellular destination, and interactions of proteins, thereby, impacting cellular physiology. Voltage-gated K+ (Kv3) channels have two conserved N-glycosylation sites which are vital for Kv3 channel activity, and primary motor neuron development. Our previous studies showed that the parental (NB_1) and N-glycan mutant (NB_1(-Mgat1), NB_1(-Mgat2), and NB_1(-Mgat3)) Neuroblastoma (NB) cell lines have compromised N-acetylglucosaminyltransferase activities: GnT-I, GnT-II, or GnT-III. Herein, we stably expressed Kv3.1b in the parental and engineered N-glycan mutant Neuroblastoma (NB) cell lines to examine how changes in N-glycans alter the cytoskeleton, and subsequently cellular properties. MALDI-TOF MS verified that the parental and mutant cell lines had different N-glycan profiles. When Kv3.1b was expressed in NB cells with an intact Mgat1, the N-glycan population had more complex N-glycans with increased branching. Further NB cells with an intact Mgat2 had higher and lower levels of hybrid and oligomannose N-glycans, respectively. N-Glycan populations changed cytoskeletal protein abundancies and cell morphology. Moreover, all Kv3.1b-expressing cells, except NB_1(-Mgat2), had changed levels of F-actin, neurofilament and vimentin, along with modified neurite formation. In all cases, migratory rates were enhanced when cells expressed Wt Kv3.1b. Glycan populations and glycans attached to Kv3.1b altered spatial arrangement in membranes and both ER folding and transport proteins were not increased by expression of unglycosylated Kv3.1b. Kv3.1b expression in NB cells alters N-glycan populations and mediates adjustments in cytoskeletal proteins, and thereby cell morphology and cell migration, supporting roles in neuronal development and maintenance.
Keywords: Cell morphology; Cell motility; Cytoskeletal proteins; Glycans; Kv3 channels; N-glycosylation; Protein folding.
Figures






Similar articles
-
Membrane Distribution and Activity of a Neuronal Voltage-Gated K+ Channel is Modified by Replacement of Complex Type N-Glycans with Hybrid Type.J Glycobiol. 2017;6(3):128. doi: 10.4172/2168-958X.1000128. Epub 2017 Oct 31. J Glycobiol. 2017. PMID: 30271698 Free PMC article.
-
Limited N-Glycan Processing Impacts Chaperone Expression Patterns, Cell Growth and Cell Invasiveness in Neuroblastoma.Biology (Basel). 2023 Feb 11;12(2):293. doi: 10.3390/biology12020293. Biology (Basel). 2023. PMID: 36829569 Free PMC article.
-
Knockdown of N-Acetylglucosaminyltransferase-II Reduces Matrix Metalloproteinase 2 Activity and Suppresses Tumorigenicity in Neuroblastoma Cell Line.Biology (Basel). 2020 Apr 4;9(4):71. doi: 10.3390/biology9040071. Biology (Basel). 2020. PMID: 32260356 Free PMC article.
-
Regulation of intracellular activity of N-glycan branching enzymes in mammals.J Biol Chem. 2024 Jul;300(7):107471. doi: 10.1016/j.jbc.2024.107471. Epub 2024 Jun 13. J Biol Chem. 2024. PMID: 38879010 Free PMC article. Review.
-
Protein glycosylation in cancers and its potential therapeutic applications in neuroblastoma.J Hematol Oncol. 2016 Sep 29;9(1):100. doi: 10.1186/s13045-016-0334-6. J Hematol Oncol. 2016. PMID: 27686492 Free PMC article. Review.
References
-
- Stanley P, Taniguchi N, Aebi M (2015) N-Glycans. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, et al. (Eds.), (3rd edn), Essentials of Glycobiology, Cold Spring Harbor Laboratory Press, New York, USA, pp. 99–111.
-
- Grissmer S, Ghanshani S, Dethlefs B, McPherson JD, Wasmuth JJ, et al. (1992) The Shaw-related potassium channel gene, Kv3.1, on human chromosome 11, encodes the type l K+ channel in T cells. J Biol Chem 267(29): 20971–20979. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
