Evaluation of lncRNAs as Potential Biomarkers for Diagnosis of Metastatic Triple-Negative Breast Cancer through Bioinformatics and Machine Learning
- PMID: 39737204
- PMCID: PMC11682528
- DOI: 10.30498/ijb.2024.432171.3853
Evaluation of lncRNAs as Potential Biomarkers for Diagnosis of Metastatic Triple-Negative Breast Cancer through Bioinformatics and Machine Learning
Abstract
Background: Triple-negative breast cancer (TNBC) is highly invasive and metastatic to the lymph nodes. Therefore, it is an urgent priority to distinguish novel biomarkers and molecular mechanisms of lymph node metastasis as the first step to the disease investigation. Long non-coding RNAs (lncRNAs) have widely been explored in cancer tumorigenesis, progression, and invasion.
Objectives: This study aimed to identify and evaluate lncRNAs in the signaling pathway of MMP11 gene in both metastatic and non-metastatic TNBC samples. The potential of lncRNAs in prognosis and diagnosis of the disease was also assessed using bioinformatics analysis, machine learning, and quantitative real-time PCR.
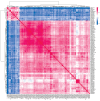
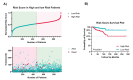
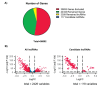
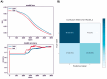
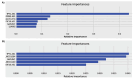
Materials and methods: Using machine learning algorithms, we analyzed the available BC data from the Cancer Genome Atlas Network (TCGA) and identified three potential lncRNAs, gastric adenocarcinoma-associated, positive CD44 regulator, long intergenic noncoding RNA (GAPLINC), TPT1-AS1, and EIF1B antisense RNA 1 (EIF1B-AS1) that could successfully distinguish between metastatic and non-metastatic TNBC.
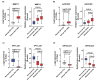
Results: The results showed the upregulation of GAPLINC lncRNA in metastatic BC tissues, compared to non-metastatic (P<0.01) and normal samples, though TPT1-AS1 and EIF1B-AS1 were downregulated in metastatic TNBC samples (P<0.01).
Conclusion: Given the aberrant expression of candidate lncRNAs and the underlying mechanisms, the above-mentioned RNAs could act as novel diagnostic and prognostic biomarkers in metastatic BC.
Keywords: Biomarkers; Breast neoplasms; Long noncoding RNA; Neoplasm metastasis; Triple negative breast neoplasms.
Copyright: © 2021 The Author(s); Published by Iranian Journal of Biotechnology.
Figures
References
LinkOut - more resources
Full Text Sources
Miscellaneous
