Flow-cytometry reveals mitochondrial DNA accumulation in Saccharomyces cerevisiae cells during cell cycle arrest
- PMID: 39737342
- PMCID: PMC11683134
- DOI: 10.3389/fcell.2024.1497652
Flow-cytometry reveals mitochondrial DNA accumulation in Saccharomyces cerevisiae cells during cell cycle arrest
Abstract
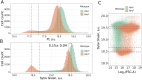
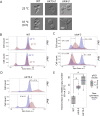
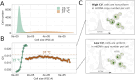
Mitochondria are semi-autonomous organelles containing their own DNA (mtDNA), which is replicated independently of nuclear DNA (nDNA). While cell cycle arrest halts nDNA replication, mtDNA replication continues. In Saccharomyces cerevisiae, flow cytometry enables semi-quantitative estimation of mtDNA levels by measuring the difference in signals between cells lacking mtDNA and those containing mtDNA. In this study, we used flow cytometry to investigate mtDNA accumulation in yeast cells under G1 and G2 phase cell cycle arrest conditions utilising thermosensitive mutants cdc4-3 and cdc15-2. In line with the previous studies, cell cycle arrest induced a several-fold accumulation of mtDNA in both mutants. The total DNA levels in arrested cells correlated with cell forward scattering, suggesting a relationship between individual cell mtDNA quantity and size. In cell cycle-arrested cells, we observed no correlation between cell size and intercellular mtDNA copy number variability. This implies that as cell size increases during arrest, the mtDNA content remains within a specific limited range for each size class. This observation suggests that mtDNA quantity control mechanisms can function in cell cycle-arrested cells.
Keywords: cell cycle arrest; cell cycle defect; mtDNA; mtDNA copy number; mtDNA copy number control; yeast.
Copyright © 2024 Potapenko, Kashko and Knorre.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Ameijeiras-Alonso J., Crujeiras R. M., Rodriguez-Casal A. (2021). Multimode: an R package for mode assessment. J. Stat. Softw. 97, 1–32. 10.18637/jss.v097.i09 - DOI
LinkOut - more resources
Full Text Sources

