Meta-QTL mapping for wheat thousand kernel weight
- PMID: 39737382
- PMCID: PMC11682887
- DOI: 10.3389/fpls.2024.1499055
Meta-QTL mapping for wheat thousand kernel weight
Abstract
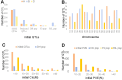
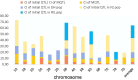
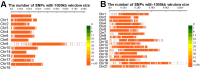
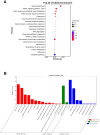
Wheat domestication and subsequent genetic improvement have yielded cultivated species with larger seeds compared to wild ancestors. Increasing thousand kernel weight (TKW) remains a crucial goal in many wheat breeding programs. To identify genomic regions influencing TKW across diverse genetic populations, we performed a comprehensive meta-analysis of quantitative trait loci (MQTL), integrating 993 initial QTL from 120 independent mapping studies over recent decades. We refined 242 loci into 66 MQTL, with an average confidence interval (CI) 3.06 times smaller than that of the original QTL. In these 66 MQTL regions, a total of 4,913 candidate genes related to TKW were identified, involved in ubiquitination, phytohormones, G-proteins, photosynthesis, and microRNAs. Expression analysis of the candidate genes showed that 95 were specific to grain and might potentially affect TKW at different seed development stages. These findings enhance our understanding of the genetic factors associated with TKW in wheat, providing reliable MQTL and potential candidate genes for genetic improvement of this trait.
Keywords: QTL mapping; genetic populations; meta-analysis; thousand kernel weight; wheat.
Copyright © 2024 Tan, Guo, Dong, Li, Chen, Cheng, Pu, Yuan and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Badji A., Otim M., Machida L., Odong T., Kwemoi D. B., Okii D., et al. (2018). Maize Combined insect resistance genomic regions and their co-localization with cell wall constituents revealed by tissue-speific QTL meta-analyses. Front. Plant Sci. 9, 895–912. doi: 10.3390/fpls.2018.00895 - DOI - PMC - PubMed
-
- Bilgrami S. S., Ramandi H. D., Shariati V., Razavi K., Tavakol E., Fakheri B. A., et al. (2020). Detection of genomic regions associated with tiller number in Iranian bread wheat under different water regimes using genome-wide association study. Sci. Rep. 10, 14034–14051. doi: 10.1038/s41598-020-69442-9 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

