Transcriptomic analysis of ROS1+ non-small cell lung cancer reveals an upregulation of nucleotide synthesis and cell adhesion pathways
- PMID: 39737401
- PMCID: PMC11683107
- DOI: 10.3389/fonc.2024.1408697
Transcriptomic analysis of ROS1+ non-small cell lung cancer reveals an upregulation of nucleotide synthesis and cell adhesion pathways
Abstract
Introduction: The transcriptomic characteristics of ROS1+ non-small cell lung cancer (NSCLC) represent a crucial aspect of its tumor biology. These features provide valuable insights into key dysregulated pathways, potentially leading to the discovery of novel targetable alterations or biomarkers.
Methods: From The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) databases, all available ROS1+ (n = 10), ALK+ (n = 5) and RET+ (n = 5) NSCLC tumor and ROS1+ cell line (n = 7) RNA-sequencing files were collected. In addition, 10 healthy lung RNA-seq samples were included. Differential gene expression with DESeq2 (R package) and gene co-expression (WGCNA, R package) analyses were performed. Functional annotation was performed through Gene Set Enrichment Analysis (GSEA) using Webgestalt and RNAseqChef, Over-Representation Analysis (ORA) through Enrichr. iRegulon was used to identify enriched transcription factors that regulate a gene co-expression module.
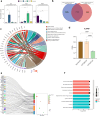
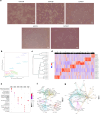
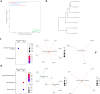
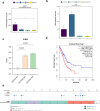
Results: ROS1+ NSCLC samples were significantly enriched for the nucleotide synthesis and cell adhesion KEGG pathways compared to ALK+ and RET+ samples. Moreover, NOTCH1 was significantly downregulated in ROS1+ NSCLC and PD-L1 was weakly expressed. When comparing ROS1+ tumor versus cell line transcriptomes, an upregulation of MYC and MET was found in cell lines together with a significantly decreased expression of HER3, HER4 and BRAF. Within ROS1-tumors, GJB2 was overexpressed in the CD74- and CLTC-ROS1+ subgroups. The differential expression of IL20RB and GJB2 in cell lines was confirmed through RT-qPCR. Finally, the gene co-expression analysis unveils a gene cluster involving cell cycle-related genes which significantly correlates with the disease stage of patients. In addition, we propose TFDP1 and ISL1 as key ROS1-specific transcription factors.
Conclusion: This study highlights cell adhesion and nucleotide synthesis as crucial signatures in ROS1+ NSCLC. The upregulation of GJB2 may serve as a prognostic biomarker, along with IL20RB, a known mediator of bone metastases. Furthermore, TDFP1 and ISL1 were identified as relevant transcription factors that could potentially regulate the biological processes in ROS1-rearranged NSCLC.
Keywords: RNA-sequencing; ROS1+ NSCLC; cell adhesion; gene co-expression; nucleotide synthesis; prognostic biomarker.
Copyright © 2024 Terrones, Op de Beeck, Van Camp, Vandeweyer and Mateiu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

