Deep Learning-Based Ion Channel Kinetics Analysis for Automated Patch Clamp Recording
- PMID: 39737527
- PMCID: PMC12083860
- DOI: 10.1002/advs.202404166
Deep Learning-Based Ion Channel Kinetics Analysis for Automated Patch Clamp Recording
Abstract
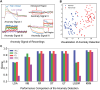
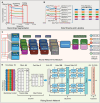
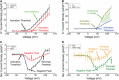
The patch clamp technique is a fundamental tool for investigating ion channel dynamics and electrophysiological properties. This study proposes the first artificial intelligence framework for characterizing multiple ion channel kinetics of whole-cell recordings. The framework integrates machine learning for anomaly detection and deep learning for multi-class classification. The anomaly detection excludes recordings that are incompatible with ion channel behavior. The multi-class classification combined a 1D convolutional neural network, bidirectional long short-term memory, and an attention mechanism to capture the spatiotemporal patterns of the recordings. The framework achieves an accuracy of 97.58% in classifying 124 test datasets into six categories based on ion channel kinetics. The utility of the novel framework is demonstrated in two applications: Alzheimer's disease drug screening and nanomatrix-induced neuronal differentiation. In drug screening, the framework illustrates the inhibitory effects of memantine on endogenous channels, and antagonistic interactions among potassium, magnesium, and calcium ion channels. For nanomatrix-induced differentiation, the classifier indicates the effects of differentiation conditions on sodium and potassium channels associated with action potentials, validating the functional properties of differentiated neurons for Parkinson's disease treatment. The proposed framework is promising for enhancing the efficiency and accuracy of ion channel kinetics analysis in electrophysiological research.
Keywords: deep learning; electrophysiology; ion channels; patch clamp; whole‐cell recording.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Application of Machine-Learning Methods to Recognize mitoBK Channels from Different Cell Types Based on the Experimental Patch-Clamp Results.Int J Mol Sci. 2021 Jan 15;22(2):840. doi: 10.3390/ijms22020840. Int J Mol Sci. 2021. PMID: 33467711 Free PMC article.
-
Utilising Automated Electrophysiological Platforms in Epilepsy Research.Methods Mol Biol. 2021;2188:133-155. doi: 10.1007/978-1-0716-0818-0_7. Methods Mol Biol. 2021. PMID: 33119850
-
Recording of multiple ion current components and action potentials in human induced pluripotent stem cell-derived cardiomyocytes via automated patch-clamp.J Pharmacol Toxicol Methods. 2019 Nov-Dec;100:106599. doi: 10.1016/j.vascn.2019.106599. Epub 2019 Jun 20. J Pharmacol Toxicol Methods. 2019. PMID: 31228558
-
The evolution of patch-clamp electrophysiology: Robotic, multiplex, and dynamic.Mol Pharmacol. 2025 Jan;107(1):100001. doi: 10.1124/molpharm.124.000954. Epub 2024 Nov 22. Mol Pharmacol. 2025. PMID: 39919159 Review.
-
An Introduction to Patch Clamp Recording.Methods Mol Biol. 2021;2188:1-19. doi: 10.1007/978-1-0716-0818-0_1. Methods Mol Biol. 2021. PMID: 33119844 Review.
Cited by
-
Deep Learning-Based Classification of Peptide Analytes from Single-Channel Nanopore Translocation Events.bioRxiv [Preprint]. 2025 May 6:2025.05.04.652126. doi: 10.1101/2025.05.04.652126. bioRxiv. 2025. PMID: 40654724 Free PMC article. Preprint.
-
Harnessing artificial intelligence for brain disease: advances in diagnosis, drug discovery, and closed-loop therapeutics.Front Neurol. 2025 Jul 28;16:1615523. doi: 10.3389/fneur.2025.1615523. eCollection 2025. Front Neurol. 2025. PMID: 40791911 Free PMC article. Review.
References
-
- Vandael D., Okamoto Y., Borges‐Merjane C., Vargas‐Barroso V., Suter B. A., Jonas P., Nat. Protoc. 2021, 16, 2947. - PubMed
-
- Minakuchi T., Guthman E. M., Acharya P., Hinson J., Fleming W., Witten I. B., Oline S. N., Falkner A. L., Nat. Neurosci. 2024, 24, 702. - PubMed
-
- Choi S. B., Polter A. M., Nemes P., Anal. Chem. 2021, 94, 1637. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
