Machine learning-enabled virtual screening indicates the anti-tuberculosis activity of aldoxorubicin and quarfloxin with verification by molecular docking, molecular dynamics simulations, and biological evaluations
- PMID: 39737570
- PMCID: PMC11684895
- DOI: 10.1093/bib/bbae696
Machine learning-enabled virtual screening indicates the anti-tuberculosis activity of aldoxorubicin and quarfloxin with verification by molecular docking, molecular dynamics simulations, and biological evaluations
Abstract
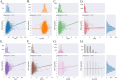
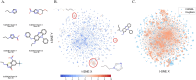
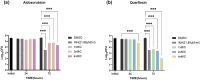
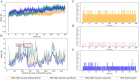
Drug resistance in Mycobacterium tuberculosis (Mtb) is a significant challenge in the control and treatment of tuberculosis, making efforts to combat the spread of this global health burden more difficult. To accelerate anti-tuberculosis drug discovery, repurposing clinically approved or investigational drugs for the treatment of tuberculosis by computational methods has become an attractive strategy. In this study, we developed a virtual screening workflow that combines multiple machine learning and deep learning models, and 11 576 compounds extracted from the DrugBank database were screened against Mtb. Our screening method produced satisfactory predictions on three data-splitting settings, with the top predicted bioactive compounds all known antibacterial or anti-TB drugs. To further identify and evaluate drugs with repurposing potential in TB therapy, 15 screened potential compounds were selected for subsequent computational and experimental evaluations, out of which aldoxorubicin and quarfloxin showed potent inhibition of Mtb strain H37Rv, with minimal inhibitory concentrations of 4.16 and 20.67 μM/mL, respectively. More inspiringly, these two compounds also showed antibacterial activity against multidrug-resistant TB isolates and exhibited strong antimicrobial activity against Mtb. Furthermore, molecular docking, molecular dynamics simulation, and the surface plasmon resonance experiments validated the direct binding of the two compounds to Mtb DNA gyrase. In summary, our effective comprehensive virtual screening workflow successfully repurposed two novel drugs (aldoxorubicin and quarfloxin) as promising anti-Mtb candidates. The verification results provide useful information for the further development and clinical verification of anti-TB drugs.
Keywords: Mycobacterium tuberculosis; antitubercular; drug repurposing; ligand-based virtual screening; machine learning.
© The Author(s) 2024. Published by Oxford University Press.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

