R3Design: deep tertiary structure-based RNA sequence design and beyond
- PMID: 39737572
- PMCID: PMC11685104
- DOI: 10.1093/bib/bbae682
R3Design: deep tertiary structure-based RNA sequence design and beyond
Abstract
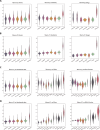
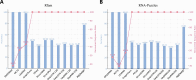
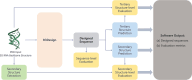
The rational design of Ribonucleic acid (RNA) molecules is crucial for advancing therapeutic applications, synthetic biology, and understanding the fundamental principles of life. Traditional RNA design methods have predominantly focused on secondary structure-based sequence design, often neglecting the intricate and essential tertiary interactions. We introduce R3Design, a tertiary structure-based RNA sequence design method that shifts the paradigm to prioritize tertiary structure in the RNA sequence design. R3Design significantly enhances sequence design on native RNA backbones, achieving high sequence recovery and Macro-F1 score, and outperforming traditional secondary structure-based approaches by substantial margins. We demonstrate that R3Design can design RNA sequences that fold into the desired tertiary structures by validating these predictions using advanced structure prediction models. This method, which is available through standalone software, provides a comprehensive toolkit for designing, folding, and evaluating RNA at the tertiary level. Our findings demonstrate R3Design's superior capability in designing RNA sequences, which achieves around $44\%$ in terms of both recovery score and Macro-F1 score in multiple datasets. This not only denotes the accuracy and fairness of the model but also underscores its potential to drive forward the development of innovative RNA-based therapeutics and to deepen our understanding of RNA biology.
Keywords: RNA; artificial intelligence; biomolecular engineering; graph neural networks; inverse folding.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures


Similar articles
-
RiboDiffusion: tertiary structure-based RNA inverse folding with generative diffusion models.Bioinformatics. 2024 Jun 28;40(Suppl 1):i347-i356. doi: 10.1093/bioinformatics/btae259. Bioinformatics. 2024. PMID: 38940178 Free PMC article.
-
Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review.
-
Frnakenstein: multiple target inverse RNA folding.BMC Bioinformatics. 2012 Oct 9;13:260. doi: 10.1186/1471-2105-13-260. BMC Bioinformatics. 2012. PMID: 23043260 Free PMC article.
-
Toward Increasing the Credibility of RNA Design.Methods Mol Biol. 2025;2847:137-151. doi: 10.1007/978-1-0716-4079-1_9. Methods Mol Biol. 2025. PMID: 39312141
-
How to benchmark RNA secondary structure prediction accuracy.Methods. 2019 Jun 1;162-163:60-67. doi: 10.1016/j.ymeth.2019.04.003. Epub 2019 Apr 2. Methods. 2019. PMID: 30951834 Free PMC article. Review.
Cited by
-
Comprehensive datasets for RNA design, machine learning, and beyond.Sci Rep. 2025 Jul 1;15(1):21417. doi: 10.1038/s41598-025-07041-2. Sci Rep. 2025. PMID: 40594473 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

