Antibodies to the RBD of SARS-CoV-2 spike mediate productive infection of primary human macrophages
- PMID: 39737903
- PMCID: PMC11686093
- DOI: 10.1038/s41467-024-54458-w
Antibodies to the RBD of SARS-CoV-2 spike mediate productive infection of primary human macrophages
Abstract
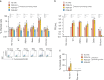
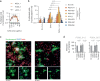
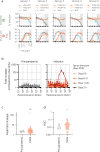
The role of myeloid cells in the pathogenesis of SARS-CoV-2 is well established, in particular as drivers of cytokine production and systemic inflammation characteristic of severe COVID-19. However, the potential for myeloid cells to act as bona fide targets of productive SARS-CoV-2 infection, and the specifics of entry, remain unclear. Using a panel of anti-SARS-CoV-2 monoclonal antibodies (mAbs) we performed a detailed assessment of antibody-mediated infection of monocytes/macrophages. mAbs with the most consistent potential to mediate infection were those targeting a conserved region of the receptor binding domain (RBD; group 1/class 4). Infection was closely related to the neutralising concentration of the mAbs, with peak infection occurring below the IC50, while pre-treating cells with remdesivir or FcγRI-blocking antibodies inhibited infection. Studies performed in primary macrophages demonstrated high-level and productive infection, with infected macrophages appearing multinucleated and syncytial. Infection was not seen in the absence of antibody with the same quantity of virus. Addition of ruxolitinib significantly increased infection, indicating restraint of infection through innate immune mechanisms rather than entry. High-level production of pro-inflammatory cytokines directly correlated with macrophage infection levels. We hypothesise that infection via antibody-FcR interactions could contribute to pathogenesis in primary infection, systemic virus spread or persistent infection.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

