Zoonotic transmission of novel Influenza A variant viruses detected in Brazil during 2020 to 2023
- PMID: 39737909
- PMCID: PMC11685428
- DOI: 10.1038/s41467-024-53815-z
Zoonotic transmission of novel Influenza A variant viruses detected in Brazil during 2020 to 2023
Abstract
Zoonotic infections (swine-human) caused by influenza A viruses (IAVs) have been reported and linked to close contact between these species. Here, we describe eight human IAV variant infections (6 mild and 2 severe cases, including 1 death) detected in Paraná, Brazil, during 2020-2023. Genomes recovered were closely related to Brazilian swIAVs of three major lineages (1 A.3.3.2/pdm09, 1B/human-like, and H3.1990.5), including three H1N1v, two H1N2v, two H3N2v and one H1v. Five H1v were closely related to pdm09 lineage, one H1v (H1N2v) grouped within 1B.2.3 clade, and the two H3v grouped within a clade composed exclusively of Brazilian H3 swIAV (clade H3.1990.5.1). Internal gene segments were closely related to H1N1pdm09 isolated from pigs. IAV variant rarely result in sustained transmission between people, however the potential to develop such ability is of concern and must not be underestimated. This study brings into focus the need for continuous influenza surveillance and timely risk assessment.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
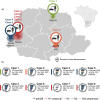





References
-
- Shinde, V. et al. Triple-Reassortant Swine Influenza A (H1) in Humans in the United States, 2005–2009. N. Engl. J. Med.360, 2616–2625 (2009). - PubMed
-
- Trifonov, V., Khiabanian, H. & Rabadan, R. Geographic Dependence, Surveillance, and Origins of the 2009 Influenza A (H1N1) Virus. N. Engl. J. Med.361, 115–119 (2009). - PubMed
-
- Trifonov, V., Khiabanian, H., Greenbaum, B. & Rabadan, R. The origin of the recent swine influenza A(H1N1) virus infecting humans. Eur. Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull.14, 19193 (2009). - PubMed
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical

